

A Novel Method for Relative Quantitation of N-Glycans via Acetone Stable Isotopic Labeling and ESI-MS Analysis
Received date: 2013-11-06
Online published: 2014-01-05
Supported by
Project supported by the National Natural Science Foundation of China (Nos. 31170773, 21375103, 31370804).
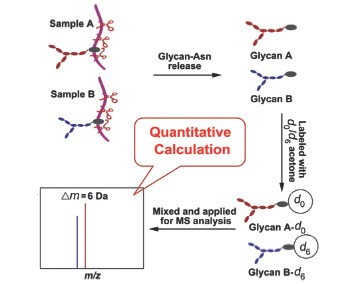
A new method for relative quantitation of N-glycans based on acetone stable isotopic labeling and ESI-MS analysis is reported. Unlike the traditional PNGase F enzymolysis method, a non-specific protease, pronase E, is employed to digest N-glycoproteins into free N-glycans bearing a single asparagine residue (N-glycan-Asns) in this study. Consequently, the Asn moiety of the obtained N-glycan-Asns exposes an active amino group for further derivatization, enabling introduction of several relative quantitation methods of proteins/peptides into glycan analysis. On this basis, the N-glycan-Asns were derivatized with acetone (d0-acetone) and its stable isotope form (d6-acetone) by reductive alkylation, which has been extensively employed for derivatization of proteins/peptides. As a result, the isotopically labeled N-glycan-Asns have the same ionization efficiency as their isotopic forms, and each N-glycan-Asn exhibits a pair of ion peaks with a stable mass difference (Dm=6 Da) in the mass spectrum. Hence, comparing the intensities of the two peaks of each glycan, the relative contents of N-glycans obtained from different glycoprotein samples can be quantified. In this work, ribonuclease B from bovine pancreas (Ribo B) was digested by pronase E and applied to ESI-MS analysis, and four peaks corresponding to high-mannose N-glycans were observed in the mass spectra. The molecular weights of these glycans were 114 Da bigger than those released by PNGase F from Ribo B, due to the persistence of the Asn residue on the pronase-obtained glycans, indicating that pronase E could stably hydrolyze the N-glycoprotein to N-glycan-Asn. Next, two same-quantity aliquots of the obtained N-glycan-Asn sample were labeled with d0-and d6-acetone, respectively, and mixed together for ESI-MS analysis. As a result, some doublet ion peaks with a 6-Da mass difference were observed in mass spectra, and the light and heavy forms of each glycan appeared almost with the same peak intensity, indicating the acetone stable isotopic labeling procedure could be steadily employed for the relatively quantitative analysis of N-glycans. Furthermore, the linearity and dynamic range of this N-glycan relative quantitation method were evaluated using mixtures of the d0-and d6-acetone labeled Ribo B N-glycan-Asn samples (Man5~Man8-Asn) in different molar ratios. The MS profiling results demonstrated that the relative quantitative method could provide the relative quantitation data with a dynamic range of 10-fold, adequate linearity (R>0.995) and reliable reproducibility with a coefficient of variation that was less than 8.7%. Moreover, the reliable relative quantitative method was successfully employed to compare the quantitative difference between different chicken egg albumin samples with different purities by MS analysis. The results showed that the relative quantitative method could accurately perform relative quantitative analysis of N-glycans.
Xue Xiangdong , Sun Yujiao , Liu Yang , Zhang Ping , Wang Zhongfu , Huang Linjuan . A Novel Method for Relative Quantitation of N-Glycans via Acetone Stable Isotopic Labeling and ESI-MS Analysis[J]. Acta Chimica Sinica, 2014 , 72(2) : 220 -226 . DOI: 10.6023/A13101106
[1] Apweiler, R.; Hermjakob, H.; Sharon, N. Acta Biochim. Biophys. Sin. 1999, 1473, 4.
[2] (a) Powlesland, A. S.; Quintero-Martinez, A.; Lim, P. G.; Pipirou, Z.; Taylor, M. E.; Drickamer, K. Methods Enzymol. 2010, 480, 165;
(b) Wu, A. M.; Liu, J.-H.; Gong, Y.-P.; Li, C.-C.; Chang, E.-T. FEBS Lett. 2010, 584, 3561.
[3] (a) Katsuhiro, O.; Tetsuya, K.; Takashi, N.; Seiichi, O. Int. J. Biochem. 1987, 23, 569;
(b) Wu, A. M.; Wu, J. H.; Singh, T.; Liu, J. H.; Tsai, M. S.; Gilboa-Garber, N. Biochimie 2006, 88, 1479.
[4] (a) Brockhausen, I. BBA-Gen. Subjects 1999, 1473, 67;
(b) Pinho, S.; Marcos, N. T.; Ferreira, B.; Carvalho, A. S.; Oliveira, M. J.; Santos-Silva, F.; Harduin-Lepers, A.. Reis, C. A. Cancer Lett. 2007, 249, 157;
(c) Wang, F.-L.; Cui, S.-X.; Sun, L.-P.; Qu, X.-J.; Xie, Y.-Y.; Zhou, L.; Mu, Y.-L.; Tang, W.; Wang, Y.-S. Cancer Detect. Prev. 2009, 32, 437.
[5] (a) Kim, Y.-G.; Jeong, H.-J.; Jang, K.-S.; Yang, Y.-H.; Song, Y.-S.; Chung, J.; Kim, B.-G. Anal. Biochem. 2009, 391, 151;
(b) Di Michele, M.; Marcone, S.; Cicchillitti, L.; Della Corte, A.; Ferlini, C.; Scambia, G.; Donati, M. B.; Rotilio, D. J. Proteomics 2010, 73, 879.
[6] (a) Miyoshi, E.; Shinzaki, S.; Moriwaki, K.; Matsumoto, H. Methods. Enzymol. 2010, 478, 153;
(b) Hirota, M.; Mogaki, M.; Pour, P. M.; Chaney, W. G. Exp. Mol. Pathol. 1993, 58, 169.
[7] (a) Wang, Z. F.; Zhang, Y.; Lin, X.; Huang, L. J. Acta Chim. Sinica 2007, 65, 2761. (王仲孚, 张英, 林雪, 黄琳娟, 化学学报, 2007, 65, 2761.);
(b) Qiu, J.; Zhang, Y.; Lu, H. J.; Yang, P. Y. Acta Chim. Sinica 2011, 69, 2123. (仇娟, 张莹, 陆豪杰, 杨芃原, 化学学报, 2011, 69, 2123.);
(c) Kuang, M.; Zhang, Y.; Yang, P. Y.; Lu, H. J. Acta Chim. Sinica 2013, 71, 1007. (匡敏, 张莹, 杨芃原, 陆豪杰, 化学学报, 2013, 71, 1007.)
[8] (a) Bowman, M. J.; Zaia, J. Anal. Chem. 2010, 82, 3023;
(b) Xia, B.; Feasley, C. L.; Sachdev, G. P.; Smith, D. F.; Cummings, R. D. Anal. Biochem. 2009, 387, 162;
(c) Yuan, J.; Hashii, N.; Kawasaki, N.; Itoh, S.; Kawanishi, T.; Hayakawa, T. J. Chromatogr. A 2005, 1067, 145;
(d) Prien, J. M.; Prater, B. D.; Qin, Q.; Cockrill, S. L. Anal. Chem. 2010, 82, 1498;
(e) Botelho, J. C.; Atwood Iii, J. A.; Cheng, L.; Alvarez-Manilla, G.; York, W. S.; Orlando, R. Int. J. Mass Spectrom. 2008, 278, 137;
(f) Zhang, P.; Zhang, Y.; Xue, X.; Wang, C.; Wang, Z.; Huang, L. Anal. Biochem. 2011, 418, 1;
(g) Bowman, M. J.; Zaia, J. Anal. Chem. 2007, 79, 5777;
(h) Alvarez-Manilla, G.; Warren, N. L.; Abney, T.; Atwood, J.; Azadi, P.; York, W. S.; Pierce, M.; Orlando, R. Glycobiology 2007, 17, 677.
[9] (a) Flory, M. R.; Griffin, T. J.; Martin, D.; Aebersold, R. Trends Biotech. 2002, 20, S23;
(b) Goshe, M. B.; Smith, R. D. Curr. Opin. Biotech. 2003, 14, 101;
(c) Gouw, J. W.; Krijgsveld, J.; Heck, A. J. R. Mol. Cell. Proteomics 2010, 9, 11;
(d) Julka, S.; Regnier, F. J. Proteome Res. 2004, 3, 350.
[10] (a) Ong, S.-E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D. B.; Steen, H.; Pandey, A.; Mann, M. Mol. Cell. Proteomics 2002, 1, 376;
(b) Krüger, M.; Moser, M.; Ussar, S.; Thievessen, I.; Luber, C. A.; Forner, F.; Schmidt, S.; Zanivan, S.; Fässler, R.; Mann, M. Cell 2008, 134, 353.
[11] Gygi, S. P.; Rist, B.; Gerber, S. A.; Turecek, F.; Gelb, M. H.; Aebersold, R. Nat. Biotechnol. 1999, 17, 994.
[12] Schmidt, A.; Kellermann, J.; Lottspeich, F. Proteomics 2005, 5, 4.
[13] Ross, P. L.; Huang, Y. N.; Marchese, J. N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; Purkayastha, S.; Juhasz, P.; Martin, S.; Bartlet-Jones, M.; He, F.; Jacobson, A.; Pappin, D. J. Mol. Cell. Proteomics 2004, 3, 1154.
[14] Liu, X.; McNally, D. J.; Nothaft, H.; Szymanski, C. M.; Brisson, J.-R.; Li, J. Anal. Chem. 2006, 78, 6081.
[15] Liu, X.; Li, X.; Chan, K.; Zou, W.; Pribil, P.; Li, X.-F.; Sawyer, M. B.; Li, J. Anal. Chem. 2007, 79, 3894.
[16] Xu, S.; Zhang, P.; Huang, L.; Wang, Z. Chem. J. Chin. Univ. 2010, 31, 1992. (徐莎, 张萍, 黄琳娟, 王仲孚, 高等学校化学学报, 2010, 31, 1992.)
[17] Zhai, J.; Liu, X.; Huang, Z.; Zhu, H. J. Am. Soc. Mass Spectr. 2009, 20, 1366.
[18] (a) Beardsley, R. L.; Karty, J. A.; Reilly, J. P. Rapid Commun. Mass Spectrom. 2000, 14, 2147;
(b) Brancia, F. L.; Oliver, S. G.; Gaskell, S. J. Rapid Commun. Mass Spectrom. 2000, 14, 2070.
[19] Fretheim, K.; Iwai, S.; Feeney, R. E. Int. J. Pept. Protein Res. 1979, 14, 451.
[20] (a) Huntington, J. A.; Stein, P. E. J. Chromatogr. B 2001, 756, 189;
(b) Charlwood, J.; Langridge, J.; Camilleri, P. Rapid Commun. Mass Spectrom. 1999, 13, 1522;
(c) Saba, J. A.; Shen, X.; Jamieson, J. C.; Perreault, H. J. Mass Spectrom. 2001, 36, 563.
[21] Packer, N. H.; Lawson, M. A.; Jardine, D. R.; Redmond, J. W. Glycoconjugate J. 1998, 15, 737.
/
| 〈 |
|
〉 |