

Whole Proteome of Fusobacterium nucleatum and Quantitative Analysis of Proteomic Change in Cancer Environment
Received date: 2015-01-06
Online published: 2015-01-29
Supported by
Project supported by MOST 863 Program (No. 2012AA020201) and CERS-1-66 (CERS-China Equipment and Education Resources System) and Research Fund for the Doctoral Program of Higher Education of China (No. 20130071140007).
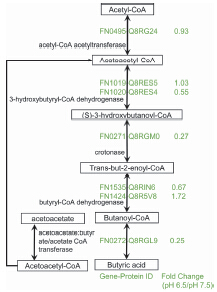
In this paper, we systematically analyzed the proteome of F. nucleatum by using liquid chromatography coupled tandem mass spectrometry (LC-MS/MS) and label free quantification in different culturing pH conditions. We identified over 950 proteins in each LC-MS/MS run. Totally 1198 proteins were identified in two different pH conditions, which correspond to 58% of the protein coding genes among the whole genome of F. nucleatum. The three replicates of pH 6.5 (pH condition of cancer tissue) and pH 7.5 (pH condition of normal tissue) coincide for more than 70% of all the identified peptides and more than 90% of the identified proteins, which indicates the stability and reproducibility of the experiments. By label-free quantitative analysis of the proteome, 1001 proteins have quantitative information. We found that 263 proteins were shown significant expression changed between pH 6.5 and pH 7.5. According to the gene ontology analysis, the significant expression changed proteins were focused on several metabolism processes and functions, such as metal ions transport, thiamin metabolic and carbohydrate metabolism and so on. Butyric acid fermentation is one of the important metabolism pathway in F. nucleatum which also have relationship with colorectal cancer. Butyric acid fermentation pathway including five categories of metabolic enzymes which contains eight protein coding genes. We have found five metabolic enzyme (acetyl-CoA acetyltransferase, 3-hydroxybutyryl-CoA dehydrogenase, crotonase, butyryl-CoA dehydrogenase and acetoacetate:butyrate/acetate CoA transferase) significant down regulation in pH condition of cancer tissue. Therefore, the butyric acid fermentation pathway were inhibited in F. nucleatum when it is living in colorectal cancer micro-environment. As the decrease of the butyric acid production is one of the phenomenon in colorectal cancer patient and F. nucleatum could promote colorectal cancer development, we verified F. nucleatum have contribution to this phenomenon. And the inhibition of butyric acid fermentation may give evidence that F. nucleatum could promote the colorectal cancer development in metabolism level.
Yin Xuefei , Liu Xiaohui , Shen Huali , Jin Hong , Yang Pengyuan . Whole Proteome of Fusobacterium nucleatum and Quantitative Analysis of Proteomic Change in Cancer Environment[J]. Acta Chimica Sinica, 2015 , 73(4) : 337 -342 . DOI: 10.6023/A15010015
[1] Group, N. H. W.; Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J. A.; Bonazzi, V.; McEwen, J. E.; Wetterstrand, K. A.; Deal, C.; Baker, C. C.; Di Francesco, V.; Howcroft, T. K.; Karp, R. W.; Lunsford, R. D.; Wellington, C. R.; Belachew, T.; Wright, M.; Giblin, C.; David, H.; Mills, M.; Salomon, R.; Mullins, C.; Akolkar, B.; Begg, L.; Davis, C.; Grandison, L.; Humble, M.; Khalsa, J.; Little, A. R.; Peavy, H.; Pontzer, C.; Portnoy, M.; Sayre, M. H.; Starke-Reed, P.; Zakhari, S.; Read, J.; Watson, B.; Guyer, M. Genome Res. 2009, 19, 2317.
[2] Backhed, F.; Ding, H.; Wang, T.; Hooper, L. V.; Koh, G. Y.; Nagy, A.; Semenkovich, C. F.; Gordon, J. I. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15718.
[3] Sjostrom, L.; Lindroos, A. K.; Peltonen, M.; Torgerson, J.; Bouchard, C.; Carlsson, B.; Dahlgren, S.; Larsson, B.; Narbro, K.; Sjostrom, C. D.; Sullivan, M.; Wedel, H.; Swedish Obese Subjects Study Scientific, G. N. Engl. J. Med. 2004, 351, 2683.
[4] Tremaroli, V.; Backhed, F. Nature 2012, 489, 242.
[5] Bolstad, A. I.; Jensen, H. B.; Bakken, V. Clin. Microbiol. Rev. 1996, 9, 55.
[6] Bennett, K. W.; Eley, A. J. Med. Microbiol. 1993, 39, 246.
[7] Kapatral, V.; Anderson, I.; Ivanova, N.; Reznik, G.; Los, T.; Lykidis, A.; Bhattacharyya, A.; Bartman, A.; Gardner, W.; Grechkin, G.; Zhu, L.; Vasieva, O.; Chu, L.; Kogan, Y.; Chaga, O.; Goltsman, E.; Bernal, A.; Larsen, N.; D'Souza, M.; Walunas, T.; Pusch, G.; Haselkorn, R.; Fonstein, M.; Kyrpides, N.; Overbeek, R. J. Bacteriol. 2002, 184, 2005.
[8] Kostic, A. D.; Gevers, D.; Pedamallu, C. S.; Michaud, M.; Duke, F.; Earl, A. M.; Ojesina, A. I.; Jung, J.; Bass, A. J.; Tabernero, J.; Baselga, J.; Liu, C.; Shivdasani, R. A.; Ogino, S.; Birren, B. W.; Huttenhower, C.; Garrett, W. S.; Meyerson, M. Genome Res. 2012, 22, 292.
[9] Castellarin, M.; Warren, R. L.; Freeman, J. D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R. A.; Holt, R. A. Genome Res. 2012, 22, 299.
[10] Rubinstein, M. R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y. W. Cell Host Microbe 2013, 14, 195.
[11] Nugent, S. G.; Kumar, D.; Rampton, D. S.; Evans, D. F. Gut 2001, 48, 571.
[12] Navratilova, J.; Hankeova, T.; Benes, P.; Smarda, J. Chemotherapy 2013, 59, 112.
[13] Cardone, R. A.; Casavola, V.; Reshkin, S. J. Nat. Rev. Cancer 2005, 5, 786.
[14] Zheng, J. F.; Sun, C. Y.; Sun, L. C.; Yang, X. M.; Wang, X. Z.; Huang, Y.; Li, Y.; He, J. Q. Acta Chim. Sinica 2010, 68, 996. (郑君芳, 孙超渊, 孙丽翠, 杨晓梅, 王小柱, 黄艳, 李洋, 贺俊崎, 化学学报, 2010, 68, 996.)
[15] Xu, F. F.; Sun, S.; Liu, X. H. Acta Chim. Sinica 2006, 64, 543. (徐菲菲, 孙胜, 刘秀华, 化学学报, 2006, 64, 543.)
[16] Liu, X. H.; Yin, X. F.; Shen, H. L.; Lu, H. J.; Yang, P. Y. Sci. Sinica (Chimica) 2014, 44, 739. (刘晓慧, 殷薛飞, 申华莉, 陆豪杰, 杨芃原, 中国科学, 2014, 44, 739.)
[17] Hebert, A. S.; Richards, A. L.; Bailey, D. J.; Ulbrich, A.; Coughlin, E. E.; Westphall, M. S.; Coon, J. J. Mol. Cell. Proteomics 2014, 13, 339.
[18] Beck, M.; Schmidt, A.; Malmstroem, J.; Claassen, M.; Ori, A.; Szymborska, A.; Herzog, F.; Rinner, O.; Ellenberg, J.; Aebersold, R. Mol. Syst. Biol. 2011, 7, 549.
[19] Ohigashi, S.; Sudo, K.; Kobayashi, D.; Takahashi, O.; Takahashi, T.; Asahara, T.; Nomoto, K.; Onodera, H. Dig. Dis. Sci. 2013, 58, 1717.
[20] Pryde, S. E.; Duncan, S. H.; Hold, G. L.; Stewart, C. S.; Flint, H. J. FEMS Microbiol. Lett. 2002, 217, 133.
[21] Scharlau, D.; Borowicki, A.; Habermann, N.; Hofmann, T.; Klenow, S.; Miene, C.; Munjal, U.; Stein, K.; Glei, M. Mutat. Res. 2009, 682, 39.
[22] Han, Y. W. Curr. Protoc. Microbiol. 2006, Chapter 13, Unit 13A 1
[23] .Huang, D. W.; Sherman, B. T.; Lempicki, R. A. Nature Protoc. 2009, 4, 44.
/
| 〈 |
|
〉 |