

Detection of Single Oligonucleotide by an Aerolysin Nanopore
Received date: 2016-07-19
Online published: 2016-08-10
Supported by
Project supported by the National Natural Science Foundation of China (Nos. 21327807, 21421004) and the Program of Introducing Talents of Discipline to Universities (No. B16017).
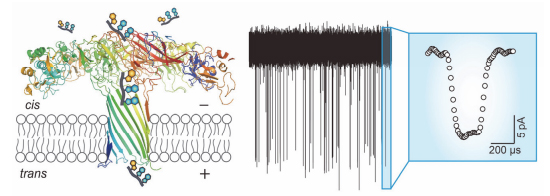
Since the nanopore single-molecule technology has been proposed, it remains a big challenge to generate a sensitive and stable nano-scale pore. In order to achieve this goal, membrane proteins, solid-state nanopore and other materials such as DNA origami have been involved to fabricate a suitable nanopore. Compared to the solid-state nanopores, biological nanopores perform a higher resolution for single molecule analysis. Therefore, the investigation of finding new biological nanopores is very important to realize the discrimination of single oligonucleotide. Aerolysin biological nanopore has been applied for the study of oligosaccharides, peptides, protein unfolding and small organic molecules so far. Here, we report that Aerolysin could be utilized for oligonucleotide analysis. The data demonstrated that Aerolysin nanopore has a high resolution both for current and time compared with other most widely used wild-type biological nanopores, such as α-hemolysin and Mycobacterium smegmatis porin A (MspA). It may be because of its narrow diameter and positive charged amino acids in the lumen. One Aerolysin pore generates a 50 pA constant ion current in 1 mol/L KCl solution, as a three nucleotides length oligonucleotides (5'-AGG-3') traversing through nanopore could induce nearly 40% current blockage. In comparison, no current blockage signals were observed when 5'-AGG-3' driven from either cis or trans side of the α-hemolysin nanopore. Furthermore, the statistical analysis of duration time of single oligonucleotide through Aerolysin indicates a relationship scale with applied voltage, as the voltage increased from 80 to 160 mV, the duration gradually decreased. Although Aerolysin nanopore has been investigated for nearly 10 years, its ability to detect oligonucleotide was not highlighted. Our findings explored high sensing capabilities of Aerolysin nanopore in the analysis of single oligonucleotide, and extended its application to single-molecule nucleic acid analysis. Aerolysin is a promising candidate for the DNA sensing, DNA damage detection, microRNA analysis and other single molecule investigations.
Cao Chan , Liao Dongfang , Ying Yilun , Long Yitao . Detection of Single Oligonucleotide by an Aerolysin Nanopore[J]. Acta Chimica Sinica, 2016 , 74(9) : 734 -737 . DOI: 10.6023/A16070352
[1] Kasianowicz, J. J.; Brandin, E.; Branton, D.; Deamer, D. W. Proc. Natl. Acad. Sci. U. S. A. 1996, 93(24), 13770.
[2] Ying, Y.-L.; Zhang, X.; Liu, Y.; Xue, M.-Z.; Li, H.-L.; Long, Y.-T. Acta Chim. Sinica 2013, 71, 44 (in Chinese). (应佚伦, 张星, 刘钰, 薛梦竹, 李洪林, 龙亿涛, 化学学报, 2013, 71, 44.)
[3] Wang, Y.; Yu, X.-F.; Liu, Y.-Y.; Xie, X.; Cheng, X.-L.; Huang, S.-M.; Wang, Z.-M. Acta Chim. Sinica 2014, 72, 378 (in Chinese). (王跃, 余旭丰, 刘芸芸, 谢骁, 程秀兰, 黄少铭, 王志民, 化学学报, 2014, 72, 378.)
[4] Ying, Y.-L.; Cao, C.; Long, Y.-T. Analyst 2014, 139, 3826.
[5] Branton, D.; Deamer, D. W.; Marziali, A.; Bayley, H.; Benner, S. A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; Jovanovich, S. B.; Krstic, P. S.; Lindsay, S.; Ling, X. S.; Mastrangelo, C. H.; Meller, A.; Oliver, J. S.; Pershin, Y. V.; Ramsey, J. M.; Riehn, R.; Soni, G. V.; Tabard-Cossa, V.; Wanunu, M.; Wiggin, M.; Schloss, J. A. Nat. Biotechnol. 2008, 26, 1146.
[6] Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Nat. Nanotechnol. 2009, 4, 265.
[7] Kumar, S.; Tao, C.; Chien, M.; Hellner, B.; Balijepalli, A.; Robertson, J. W.; Li, Z.; Russo, J. J.; Reiner, J. E.; Kasianowicz, J. J.; Ju, J. Sci. Rep. 2012, 2, 684.
[8] (a) Stoddart, D.; Heron, A. J.; Mikhailova, E.; Maglia, G.; Bayley, H. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 7702;
(b) Manrao, E. A.; Derrington, I. M.; Pavlenok, M.; Niederweis, M.; Gundlach, J. H. PLoS ONE 2011, 6, e25723.
[9] (a) Wang, H.-Y.; Li, Y.; Qin, L.-X.; Heyman, A.; Shoseyov, O.; Willner, I.; Long, Y.-T.; Tian, H. Chem. Commun. 2013, 49, 1741;
(b) Butler, T. Z.; Pavlenok, M.; Derrington, I. M.; Niederweis, M.; Gundlach, J. H. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 20647.
[10] Manrao, E. A.; Derrington, I. M.; Laszlo, A. H.; Langford, K. W.; Hopper, M. K.; Gillgren, N.; Pavlenok, M.; Niederweis, M.; Gundlach, J. H. Nat. Biotechnol. 2012, 30, 349.
[11] Parker, M. W.; Buckley, J. T.; Postma, J. P. M.; Tucker, A. D.; Leonard, K.; Pattus, F.; Tsernoglou, D. Nature 1994, 367, 292.
[12] Stefureac, R.; Long, Y.-T.; Kraatz, H.-B.; Howard, P.; Lee, J. S. Biochemistry 2006, 45, 9172.
[13] Pastoriza-Gallego, M.; Rabah, L.; Gibrat, G.; Thiebot, B.; van der Goot, F. G.; Auvray, L.; Betton, J. M.; Pelta, J. J. J. Am. Chem. Soc. 2011, 133, 2923.
[14] Baaken, G.; Halimeh, I.; Bacri, L.; Pelta, J.; Oukhaled, A.; Behrends, J. C. ACS Nano 2015, 9, 6443.
[15] Fennouri, A.; Przybylski, C.; Pastoriza-Gallego, M.; Bacri, L.; Auvray, L.; Daniel, R. ACS Nano 2012, 6, 9672.
[16] (a) Fennouri, A.; Daniel, R.; Pastoriza-Gallego, M.; Auvray, L.; Pelta, J.; Bacri, L. Anal. Chem. 2013, 85, 8488;
(b) Cressiot, B.; Braselmann, E.; Oukhaled, A.; Elcock, A. H.; Pelta, J.; Clark, P. L. ACS Nano 2015, 9, 9050.
[17] Degiacomi, M. T.; Iacovache, I.; Pernot, L.; Chami, M.; Kudryashev, M.; Stahlberg, H.; van der Goot, F. G.; Peraro, M. D. Nat. Chem. Biol. 2013, 9, 623.
[18] Chan, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Nat. Nanotechnol. 2016, 11, 713.
[19] Cao, C.; Ying, Y.-L.; Gu, Z.; Long, Y.-T. Anal. Chem. 2014, 86, 11946.
/
| 〈 |
|
〉 |