

Theoretical Insight into the Catalytic Mechanism of Enoyl-CoA Hydratase
Received date: 2016-10-20
Online published: 2017-03-21
Supported by
Project supported by the Natural Science Foundation of Zhejiang Province (LY16B070002) and Technological Innovation Plan & New Talent Plan for College Students in Zhejiang Province (2015R404006).
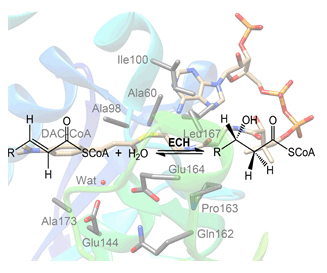
Enoyl-CoA hydratase (ECH), which is also known as crotonase, is the second requisite enzyme in the β-oxidation pathway of fatty acid that catalyzes the syn hydration of α,β-unsaturated thiolester substrates. In this work, ECH-catalyzed hydration mechanisms of DAC-CoA and Crotonyl-CoA were investigated using density functional theory (DFT) methods. Geometrical structures were optimized using Gaussian 03 program at the B3LYP/6-31G(d,p) level of theory. Frequency calculations were performed with the 6-31G(d,p) basis set to obtain zero-point vibrational energies (ZPEs) and to confirm the nature of all the stationary points that have no imaginary frequency for the local minima and have only one imaginary frequency for the saddle points. The single-point calculations on the optimized geometries were further performed with 6-311++G(2d,2p) basis set to obtain more accurate energies. The polarizable-continuum model (PCM) with the dielectric constant of 4 was used to calculate the single point energies at 6-311++G(2d, p) level on all the optimized geometries to consider the effects of enzymatic environment that was not included in the computational model. Considering that B3LYP functional lacks the proper description of the long-range dispersion interactions, we further used the DFT-D3 program to calculate the empirical dispersion correction to correct the B3LYP energies. The final energies reported in this work are the single-point energies corrected for ZPEs, solvation and dispersion effects. The calculated results suggested that hydration proceeds through a stepwise mechanism, involving an enolate intermediate. Glu164 functions as the sole base/acid for catalysis. Although Glu144 is not directly involved in hydration, it induces the catalytic water molecule to locate an ideal orientation to attack the double bond of substrate by the hydrogen-bonding interaction. Crotonyl-CoA shows higher hydration activity than DAC-CoA. The backbone NH groups of Ala98 and Gly141 form an oxyanion hole with substrate carbonyl oxygen, which play key roles in binding substrate and stabilizing the generated transition states and intermediates. In addition, the hydrogen-bonding networks surrounding Glu144 and Glu164 are of great importance for active site arrangement.
Zhang Yu , Yang Xinya , Yu Haiying , Ma Guangcai . Theoretical Insight into the Catalytic Mechanism of Enoyl-CoA Hydratase[J]. Acta Chimica Sinica, 2017 , 75(5) : 494 -500 . DOI: 10.6023/A16100559
[1] Willadsen, P.; Eggerer, H. Eur. J. Biochem. 1975, 54, 247.
[2] Wu, W. J.; Feng, Y.; He, X.; Hofstein, H. A.; Raleigh, D. P.; Tonge, P. J. J. Am. Chem. Soc. 2000, 122, 3987.
[3] Bahnson, B. J.; Anderson, V. E. Biochemistry 1989, 28, 4173.
[4] Müller-Newen, G.; Janssen, U.; Stoffel, W. Eur. J. Biochem. 1995, 228, 68.
[5] Boersma, A. J.; Coquière, D.; Geerdink, D.; Rosati, F.; Feringa, B. L.; Roelfes, G. Nat. Chem. 2010, 2, 991.
[6] Silverman, R. B. The Organic Chemistry of Enzyme-catalyzed Reactions, Academic, London, 2002, pp. 428~448.
[7] Engel, C. K.; Mathieu, M.; Zeelen, J. P.; Hiltunen, J. K.; Wierenga, R. K. EMBO J. 1996, 15, 5135.
[8] Bahnson, B. J.; Anderson, V. E.; Petsko, G. A. Biochemistry 2002, 41, 2621.
[9] Hisano, T.; Tsuge, T.; Fukui, T.; Iwata, T.; Miki, K.; Doi, Y. J. Biol. Chem. 2003, 278, 617.
[10] Koski, M. K.; Haapalainen, A. M.; Hiltunen, J. K.; Glumoff, T. J. Biol. Chem. 2004, 279, 24666.
[11] Baugh, L.; Phan, I.; Begley, D. W.; Clifton, M. C.; Armour, B.; Dranow, D. M.; Taylor, B. M.; Muruthi, M. M.; Abendroth, J.; Fairman, J. W.; Fox, D. 3rd; Dieterich, S. H.; Staker, B. L.; Gardberg, A. S.; Choi, R.; Hewitt, S. N.; Napuli, A. J.; Myers, J.; Barrett, L. K.; Zhang, Y.; Ferrell, M.; Mundt, E.; Thompkins, K.; Tran, N.; Lyons-Abbott, S.; Abramov, A.; Sekar, A.; Serbzhinskiy, D,; Lorimer, D.; Buchko, G. W.; Stacy, R.; Stewart, L. J.; Edwards, T. E.; Van Voorhis, W. C.; Myler, P. J. Tuberculosis 2015, 95, 142.
[12] Feng, Y.; Hofstein, H. A.; Zwahlen, J.; Tonge, P. J. Biochemistry 2002, 41, 12883.
[13] Hofstein, H. A.; Feng, Y.; Anderson, V. E.; Tonge, P. J. Biochemistry 1999, 38, 9508.
[14] Engel, C. K.; Kiema, T. R.; Hiltunen, J. K.; Wierenga, R. K. J. Mol. Biol. 1998, 275, 847.
[15] Bell, A. F.; Wu, J.; Feng, Y.; Tonge, P. J. Biochemistry 2001, 40, 1725.
[16] D'Ordine, R. L.; Pawlak, J.; Bahnson, B. J.; Anderson, V. E.; Biochemistry 2002, 41, 2630.
[17] Bahnson, B. J.; Anderson, V. E. Biochemistry 1991, 30, 5894.
[18] Pawlak, J.; Bahnson, B.; Anderson, V. Nukleonika 2002, 47, 33.
[19] Cui, X.; He, R.; Yang, Q.; Shen, W.; Li, M. J. Mol. Model. 2014, 20, 2411.
[20] Agnihotri, G.; Liu, H. W. Bioorg. Med. Chem. 2003, 11, 9.
[21] Siegbahn, P. E.; Himo, F. J. Biol. Inorg. Chem. 2009, 14, 643.
[22] Siegbahn, P. E.; Blomberg, M. R. Chem. Rev. 2010, 110, 7040.
[23] Hopmann, K. H.; Himo, F. In Comprehensive Natural Products Chemistry II Chemistry and Biology, Vol. 8, Eds.: Mander, L. N.; Liu, H.-W., Elsevier, Oxford, 2010, pp. 719~747.
[24] Siegbahn, P. E.; Himo, F. Wiley Interdisciplinary Reviews: Computational Molecular Science, 2011, 1, 323.
[25] Blomberg, M. R.; Borowski, T.; Himo, F.; Liao, R. Z.; Siegbahn, P. E. Chem. Rev. 2014, 114, 3601.
[26] Becke, A. D. J. Chem. Phys. 1993, 98, 5648.
[27] Lee, C.; Yang, W.; Parr, R. G. Phys. Rev. B 1988, 37, 785.
[28] Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Montgomery, J. A., Jr.; Vreven, T.; Kudin, K. N.; Burant, J. C.; Millam, J. M.; Iyengar, S. S.; Tomasi, J.; Barone, V.; Mennucci, B.; Cossi, M.; Scalmani, G.; Rega, N.; Petersson, G. A.; Nakatsuji, H.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Klene, M.; Li, X.; Knox, J. E.; Hratchian, H. P.; Cross, J. B.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R. E.; Yazyev, O.; Austin, A. J.; Cammi, R.; Pomelli, C.; Ochterski, J. W.; Ayala, P. Y.; Morokuma, K.; Voth, G. A.; Salvador, P.; Dannenberg, J. J.; Zakrzewski, V. G.; Dapprich, S.; Daniels, A. D.; Strain, M. C.; Farkas, O.; Malick, D. K.; Rabuck, A. D.; Raghavachari, K.; Foresman, J. B.; Ortiz, J. V.; Cui, Q.; Baboul, A. G.; Clifford, S.; Cioslowski, J.; Stefanov, B. B.; Liu, G.; Liashenko, A.; Piskorz, P.; Komaromi, I.; Martin, R. L.; Fox, D. J.; Keith, T.; Al-Laham, M. A.; Peng, C. Y.; Nanayakkara, A.; Challacombe, M.; Gill, P. M. W.; Johnson, B.; Chen, W.; Wong, M. W.; Gonzalez, C.; Pople, J. A. Gaussian 03 (Revision D. 01), Gaussian, Inc., Wallingford CT, 2004.
[29] Barone, V.; Cossi, M.; Tomasi, J. J. Comput. Chem. 1998, 19, 404.
[30] Tomasi, J.; Persico, M. Chem. Rev. 1994, 94, 2027.
[31] Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. J. Chem. Phys. 2010, 132, 154104.
[32] Grimme, S.; Ehrlich, S.; Goerigk, L. J. Comput. Chem. 2011, 32, 1456.
[33] D'Ordine, R. L.; Tonge, P. J.; Carey, P. R.; Anderson, V. E. Biochemistry 1994, 33, 12635.
/
| 〈 |
|
〉 |