

Spectral and Computational Simulations of HSA and BDE154 Based on Acidity Induction
Received date: 2018-02-06
Online published: 2018-04-11
Supported by
Project supported by the National Natural Science Foundation of China (Nos. 21467006 and 21565012) and the Guangxi Natural Science Foundation of China (2017GXNSFAA198354).
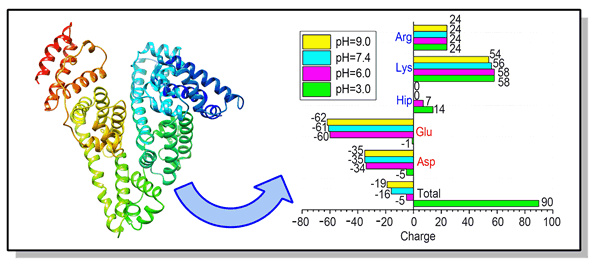
In this paper, human serum albumin (HSA) binding to small molecule 2,2',4,4',5,6'-hexabromodiphenyl ether (BDE154) is studied by means of inducing protonation or deprotonation at four different pH levels (pH=3.0, 6.0, 7.4, 9.0). Firstly, it has been indicated that the charge distribution on HSA is very uniform even after protonation of HSA at different pH levels. From this, it can be inferred that the uniform charge distribution makes the electrostatic forces between the amino acid residues of the ⅡA region of HSA aspartic acid (Asp),glutamate (Glu) and histidine (His) to gradually reach a relative equilibrium and thus stabilize the HSA conformation. The results from synchronous fluorescence spectroscopy show that BDE154 has been bind to the ⅡA region of HSA, and is more closely to tryptophan (Try), and that causes the fluorescence quenching of HSA. After that, the semi-flexible docking of HSA with BDE 154 reveals that BDE154 has a cationic-π-conjugated effect and strong hydrophobic interaction with the surrounding amino acids, such as tyrosine 150 (Tyr150), lysine 195 (Lys195), lysine 199 (Lys199), etc. Next, the dynamic and thermodynamic properties of HSA under different protonation conditions have been studied by using molecular dynamic simulation. The results of simulation also show that too much positive charge deteriorates the system stability of HSA or HSA-BDE154 complex. Then, the binding free energy of HSA-BDE154 complex under different protonation states has been predicted by MM-PBSA method, and the contribution of amino acid residues to free energy of binding has also been analyzed. In addition, lysine 199 (Lys199), leucine 238 (Leu238), arginine 257 (Arg257), alanine 261 (Ala261), and isoleucine 264 (Ile264) in the HSA, being located in the hydrophobic cavity in subdomain ⅡA, are the most important residues when binding with BDE154. Therefore, the hydrophobic interaction has been identified as the major driving force for the binding between HSA-BDE154 systems, which is consistent with the results of molecular docking and the analysis of binding free energy. Finally, the results of secondary structure analysis of molecular dynamics simulation show that the binding could promote the de-helix process of HSA by increasing the acidity in HSA-BDE154 complex system.
Xu Jie , Wei Yuchen , Wu Zhiwei , Yi Zhongsheng . Spectral and Computational Simulations of HSA and BDE154 Based on Acidity Induction[J]. Acta Chimica Sinica, 2018 , 76(5) : 408 -414 . DOI: 10.6023/A18020060
[1] Allard, J. F.; Dushek, O.; Coombs, D.; Van Der Merwe; P. A. Biophys. J. 2012, 102, 1265.
[2] Rich, R. L.; Hoth, L. R.; Geoghegan, K. F.; Brown, T. A.; LeMotte, P. K.; Simons, S. P.; Hensley, P.; Myszka, D. G. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 8562.
[3] Ballardini, R.; Balzani, V.; Credi, A.; Gandolfi, M. T.; Kotzyba-Hibert, F.; Lehn, J. M.; Prodi, L. J. Am. Chem. Soc. 1994, 116, 5741.
[4] Agasti, S. S.; Liong, M.; Tassa, C.; Chung, H. J.; Shaw, S. Y.; Lee, H.; Weissleder, R. Angew. Chem. Int. Ed. 2012, 51, 450.
[5] Gellman, S. H. Chem. Rev. 1997, 97, 1231.
[6] Berde, C. B.; Hudson, B. S.; Simoni, R. D.; Sklar, L. A. J. Biol. Chem. 1979, 254, 391.
[7] Schneider, H. J.; Yatsimirsky, A. K. Chem. Soc. Rev. 2008, 37, 263.
[8] Pal, B.; Bajpai, P. K.; Baul, T. B. Spectrochim. Acta A 2000, 56, 2453.
[9] Lyon, C. E.; Suh, E. S.; Dobson, C. M.; Hore, P. J. J. Am. Chem. Soc. 2002, 124, 13018.
[10] Cheng, L. T.; Wang, Z.; Setny, P.; Dzubiella, J.; Li, B.; McCammon, J. A. J. Chem. Phys. 2009, 131, 144102.
[11] Ma, M.; Paredes, A., Bong, D. J. Am. Chem. Soc. 2008, 130, 14456.
[12] Bader, A. N.; van Dongen, M. M.; van Lipzig, M. M.; Kool, J.; Meerman, J. H.; Ariese, F.; Gooijer, C. Chem. Res. Toxicol. 2005, 18, 1405.
[13] Colquhoun, H. M.; Zhu, Z.; Williams, D. J. Org. Lett. 2003, 5, 4353.
[14] Branco, T. J. F.; Ferreira, L. V.; do Rego, A. B.; Oliveira, A. S.; Da Silva, J. P. Photochem. Photobiol. Sci. 2006, 5, 1068.
[15] Baudry, R.; Kalchenko, O.; Dumazet-Bonnamour, I.; Vocanson, F.; Lamartine, R. J. Chromatogr. Sci. 2003, 41, 157.
[16] Xiao, Z.-Y.; Lin, R.-L.; Tao, Z.; Liu, Q.-Y.; Liu, J.-X.; Xiao, X. Org. Chem. Front. 2017, 4, 2422.
[17] Baudoin, O.; Gonnet, F.; Teulade-Fichou, M. P.; Vigneron, J. P.; Tabet, J. C., Lehn, J. M. Chem. Eur. J. 1999, 5, 2762.
[18] Kukic, P.; Nielsen; J. E. Future Med. Chem. 2010, 2, 647.
[19] Jakoby IV, M. G.; Miller, K. R.; Toner, J. J.; Bauman, A.; Cheng, L.; Li, E.; Cistola, D. P. Biochemistry 1993, 32, 872.
[20] Quan, X.; Dong, J.; Zhou, J. Acta Chim. Sinica 2014, 72, 1075. (全学波, 董佳奇, 周健, 化学学报, 2014, 72, 1075.)
[21] Li, L.; Lü, M.; Lu, K.; Liu, G.; Peng, L. Chin. J. Org. Chem. 2018, 38, 246. (李林璐, 吕名秀, 卢奎, 刘广斌, 彭露, 有机化学, 2018, 38, 246.)
[22] Linderstrøm-Lang, K. C. R. Trav. Lab. Carlsberg 1924, 15, 1.
[23] Shishkov, I. F.; Khristenko, L. V.; Rudakov, F. M.; Vilkov, L. V.; Karlov, S. S.; Zaitseva, G. S.; Samdal, S. J. Mol. Struct. 2002, 641, 199.
[24] Bhattacharya, B.; Nakka, S.; Guruprasad, L.; Samanta, A. J. Phys. Chem. B 2009, 113, 2143.
[25] Jang, H.; Hall, C. K.; Zhou, Y. Biophys. J. 2002, 83, 819.
[26] de Oliveira, C. A. F.; Guimarães, C. R. W.; Barreiro, G.; de Alencastro, R. B. Proteins 2003, 52, 483.
[27] Bolel, P.; Datta, S.; Mahapatra, N.; Halder, M. J. Phys. Chem. B 2014, 118, 26.
[28] Baler, K.; Martin, O. A.; Carignano, M. A.; Ameer, G. A.; Vila, J. A.; Szleifer, I. J. Phys. Chem. B 2014, 118, 921.
[29] Meyer, B.; Peters, T. Angew. Chem. Int. Ed. 2003, 42, 864.
[30] Karplus, M.; McCammon, J. A. Nat. Struct. Biol. 2002, 9, 646.
[31] Zhang, Y.; Wu, S.; Qin, Y.; Liu, J.; Liu, J.; Wang, Q.; Ren, F.; Zhang, H. Food Chem. 2018, 240, 1072.
[32] De Wit, C. A. Chemosphere 2002, 46, 583.
[33] Zhao, N.; Xuan, S.; Fronczek, F. R.; Smith, K. M.; Vicente, M. G. J. Org. Chem. 2017, 82, 3880.
[34] Balamurugan, R.; Stalin, A.; Ignacimuthu, S. Eur. J. Med. Chem. 2012, 47, 38.
[35] Dolinsky, T. J.; Czodrowski, P.; Li, H.; Nielsen, J. E.; Jensen, J. H.; Klebe, G.; Baker, N. A. Nucleic Acids Res. 2007, 35, 522.
[36] Li, Y.; Liu, X.; Dong, X.; Zhang, L.; Sun, Y. Langmuir 2014, 30, 8500.
[37] Aliev, A. E.; Kulke, M.; Khaneja, H. S.; Chudasama, V.; Sheppard, T. D.; Lanigan, R. M. Proteins:Structure, Function, and Bioinformatics 2014, 82, 195-215.
[38] Darden, T.; York, D.; Pedersen, L. J. Chem. Phys. 1993, 98, 10089.
[39] Ryckaert, J. P.; Ciccotti, G.; Berendsen, H. J. J. Comput. Phys. 1977, 23, 327.
[40] Sudhamalla, B.; Gokara, M.; Ahalawat, N.; Amooru, D. G.; Subramanyam, R. J. Phys. Chem. B 2010, 114, 9054.
[41] Baler, K.; Martin, O. A.; Carignano, M. A.; Ameer, G. A.; Vila, J. A.; Szleifer, I. J. Phys. Chem. B 2014, 118, 921.
[42] Jiang, Q.; Zhang, Z.; Liu, Y.; Yao, N.; Wang, J. Chin. J. Org. Chem. 2017, 37, 1814. (姜齐永, 张珠, 刘洋, 姚楠楠, 王进军, 有机化学, 2017, 37, 1814.)
[43] Lu, Z.; Qi, L.; Li, G.-X.; Li, Q.; Sun, G.-H.; Xie, R.-Z. J. Solution Chem. 2014, 43, 2010.
[44] Rehman, M. T.; Shamsi, H.; Khan, A. U. Mol. Pharm. 2014, 11, 1785.
[45] Rahman, T.; Rahmatullah, M. Bioorg. Med. Chem. Lett. 2010, 20, 537.
[46] Dong, K.; Yang, X.; Zhao, T.; Zhu, X. Mol. Simulat. 2017, 43, 599.
/
| 〈 |
|
〉 |