

Established and Optimized the Measurement of Serum Troponin I Using Liquid Chromatography Tandem Mass Spectrometry
Received date: 2019-01-18
Online published: 2019-05-08
Supported by
Project supported by the National Key Research and Development Program of China (No. 2017YFF0205401).
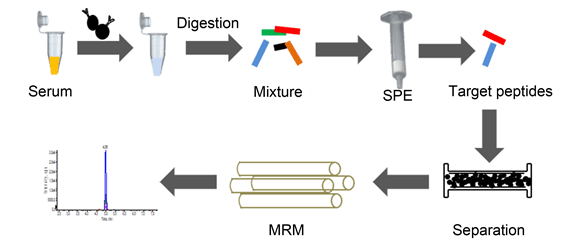
Cardiac troponin I (cTnI) is one of the most popular biomarkers for the diagnosis of acute myocardial injury (AMI) in patients. In this study, a novel method was developed and optimized for quantification of cTnI in human serum by immunoaffinity enrichment combining isotope dilution liquid chromatography tandem mass spectrometry. cTnI was first captured from human serum with immunomagnetic beads conjugated with the monoclonal antibody, and then enzymatically hydrolyzed into peptides after a series of operation, including denaturation, reduction, acetylation, digestion and purification. Subsequently, enzymatic peptides were separated by passing through Symmetry Shield C18 column at a speed of 0.2 mL/min with 0.1% acid acetonitrile solution and 0.1% acid aqueous solution as mobile phases under gradient elution. A specific peptide, NITEIADLTQK, was selected and quantified. The qualitative analysis was achieved by three ion transitions under multiple reaction monitoring (MRM) when the isotopically labeled peptide, NITEIAD[(13C6,15N)L]TQK, was used as a reference. After the optimal conditional experiment, results showed that the surrogate peptide was separated out well at about 4.95 min with little interference. Ranging from 10 ng/mL to 600 ng/mL, a good linearity was shown with correlation coefficients all above 0.99. The limit of detection (LOD) and the limit of quantification (LOQ) were estimated to be 2.5 ng/mL, 8.32 ng/mL, respectively. This method also provided good accuracy (relative bias of three concentrations diluted from SRM2921 were between -7.94% and -6.49%) and repeatability (total relative standard deviations of three concentration were 6.43%, 3.18% and 2.75%, respectively). Carry-over rates were estimated between -0.47% and 0.04%. The novel assay successfully determined five specimens from AMI patients with concentrations from 16.38 ng/mL to 557.53 ng/mL. Our results demonstrate that this method can be applied for determination of serum cTnI in AMI patients with high selectivity, low carry-over rates, good repeatability and good accuracy, which helps to establish candidate reference measurement procedure of serum cTnI.
Li Yanan , Ji Huoyan , Wang Tianyi , Shen Lei , Shi Xiuying , Wang Jianxin . Established and Optimized the Measurement of Serum Troponin I Using Liquid Chromatography Tandem Mass Spectrometry[J]. Acta Chimica Sinica, 2019 , 77(6) : 539 -544 . DOI: 10.6023/A19010032
[1] Stillman, A. E.; Oudkerk, M.; Bluemke, D.; Bremerich, J.; Esteves, F. P.; Garcia, E. V.; Gutberlet, M.; Hundley, W. G.; Jerosch-Herold, M.; Kuijpers, D.; Kwong, R. K.; Nagel, E.; Lerakis, S.; Oshinski, J.; Paul, J. F.; Underwood, R.; Witersperger, B. J.; Rees, M. R. Int. J. Cardiovasc. Imaging 2011, 27, 7.
[2] Soetkamp, D.; Raedschelders, K.; Mastali, M.; Sobhani, K.; Bairey Merz, C. N.; Van Eyk, J. Expert Rev. Proteomics 2017, 14, 973.
[3] Boriani, G.; Biffi, M.; Cervi, V.; Bronzetti, G.; Magagnoli, G.; Zannoli, R.; Branzi, A. Chest. 2000, 118, 342.
[4] Panteghini, M.; Bunk, D. M.; Christenson, R. H.; Katrukha, A.; Porter, R. A.; Schimmel, H.; Wang, L.; Tate, J. R. Clin. Chem. Lab. Med. 2008, 46, 1501.
[5] Tate, J. R.; Bunk, D. M.; Christenson, R. H.; Katrukha, A.; Noble, J. E.; Porter, R. A.; Schimmel, H.; Wang, L.; Panteghini, M. Pathology 2010, 42, 402.
[6] Hoofnagle, A. N.; Wener, M. H. J. Immunol. Methods 2009, 347, 3.
[7] Noble, J. E.; Bunk, D. M.; Christenson, R. H.; Cole, K. D.; He, H. J.; Katrukha, A. G.; Panteghini, M.; Porter, R. A.; Schimmel, H.; Tate, J. R.; Wang, L. Clin. Chem. Lab. Med. 2010, 48, 1603.
[8] Guo, Q.-Z.; Du, Z.-X. Chin. J. Chem. 2019, 29, 1922.
[9] Chen, L.-N.; Song, F.-R.; Zheng, Z.; Xing, J.-P.; Liu, Z.-Q.; Liu, S.-Y. Acta Chim. Sinica 2012, 70, 843(in Chinese). (陈丽娜, 宋凤瑞, 郑重, 邢俊鹏, 刘志强, 刘淑莹, 化学学报, 2012, 70, 843.)
[10] Agger, S. A.; Marney, L. C.; Hoofnagle, A. N. Clin. Chem. 2010, 56, 1804.
[11] Kuhn, E.; Wu, J.; Karl, J.; Liao, H.; Zolg, W.; Guild, B. Proteomics 2004, 4, 1175.
[12] Keshishian, H.; Addona, T.; Burgess, M.; Mani, D. R.; Shi, X.; Kuhn, E.; Sabatine, M. S.; Gerszten, R. E.; Carr, S. A. Mol. Cell. Proteomics 2009, 8, 2339.
[13] Kuhn, E.; Addona, T.; Keshishian, H.; Burgess, M.; Mani, D. R.; Lee, R. T.; Sabatine, M. S.; Gerszten, R. E.; Carr, S. A. Clin. Chem. 2009, 55, 1108.
[14] Li, X.-Q.; Yang, Z.; Zhang, Q.-H. J. Chin. Mass Spectrom. Soc. 2013, 34, 338(in Chinese). (李秀琴, 杨总, 张庆合, 质谱学报, 2013, 34, 338.)
[15] Anderson, N. L. Clin. Chem. 2010, 56, 177.
[16] Anderson, N. L.; Anderson, N. G. Mol. Cell. Proteomics 2002, 1, 845.
[17] Williams, D. K.; Muddiman, D. C. J. Proteome Res. 2009, 8, 1085.
[18] Ji, H.; Wang, J.; Ju, S.; Cong, H.; Wang, X.; Su, J.; Wang, H. J. Chromatogr. B:Anal. Technol. Biomed. Life Sci. 2017, 1059, 49.
[19] Mann, M.; Hendrickson, R. C.; Pandey, A. Annu. Rev. Biochem. 2001, 70, 437.
[20] Zhao, M.; Wu, F. Protein Expression Purif. 2015, 116, 120.
[21] Rice, R. H.; Means, G. E. Biochim. Biophys. Acta 1977, 492, 316.
[22] Nash, D. C.; Chase, H. A. J. Chromatogr. A 1998, 807, 185.
/
| 〈 |
|
〉 |