

Recent Advances in the Structural Studies on Cytosine Deaminase APOBEC3 Family Members and Their Nucleic Acid Complexes
Received date: 2019-08-07
Online published: 2019-08-15
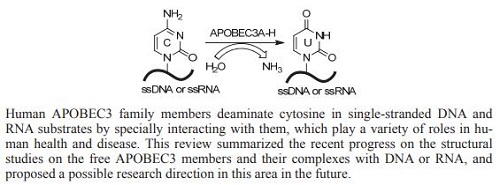
Apolipoprotein B mRNA catalytically edited protein APOBEC3 (A3) is a family of proteins in the intracellular retrotransposon defense system, including seven members APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3DE (A3DE), APOBEC3F (A3F), APOBEC3G (A3G) and APOBEC3H (A3H) encoded in a tandem array on human chromosome 22. They deaminate cytosine in single-stranded DNA and RNA substrates, which play a variety of roles in human health and disease. Among them, A3DE, A3F, A3G and A3H restrict replication of human immunodeficiency virus-1 (HIV-1) in strains lacking the virus infectivity factor protein (Vif) by deaminating cytidine in virus cDNA. Subsequent replication of the virus cDNA generates the hallmark G-to-A hyper-mutations, causing proviral inactivation. HIV-1 develops countermeasures to antagonize this intrinsic host defense response. Its Vif protein facilitates polyubiquitination of A3 members by recruiting an E3 ubiquitin ligase complex, which results in the proteasomal degradation of A3 proteins. To better understand the deamination mechanism of A3 proteins, we here reviewed the research progress on the structures of free A3 family members and their complexes with single-stranded DNA or double-stranded RNA. It includes the structures of the apo-forms of N- and/or C-termini domains of A3A, A3B, A3C, A3F, A3G and A3H, or the chimeric forms of their functional domains, and their complexes with nucleic acids, which demonstrate the basis of how A3 proteins to identify target base cytosine in hot motifs 5'-TC or 5'-CC in DNA, and then to conduct catalytic deamination. We simply described how the key residues of A3 members are involved in DNA or RNA interactions, the common properties of their structures, and their interactions with DNA or RNA. We partially discussed the interactions between A3 proteins and Vif, therefore, this review might be helpful to rationally design anti-virus drugs to disrupt these interactions. We finally suggested the new research directions about how to make full-length A3 proteins containing N-terminal CD1 and C-terminal CD2 domains, and how to study the interactions between these full-length A3 proteins and nucleic acids through cryo-EM and other techniques.
Key words: APOBEC3; cytidine deamination; HIV virus; nucleic acid; structure
Jiaoyu Jin , Xiaoxuan Yan , Yaping Liu , Wenxian Lan , Chunxi Wang , Bin Xu , Chunyang Cao . Recent Advances in the Structural Studies on Cytosine Deaminase APOBEC3 Family Members and Their Nucleic Acid Complexes[J]. Acta Chimica Sinica, 2019 , 77(11) : 1089 -1098 . DOI: 10.6023/A19080296
| [1] | Goila-Gaur R.; Strebel K. Retrovirology 2008, 5, 51. |
| [2] | Larue R. S.; Andresdottir V.; Blanchard Y.; Conticello S. G.; Derse D.; Emerman M.; Greene W. C.; Jonsson S. R.; Landau N. R.; Lochelt M.; Malik H. S.; Malim M. H.; Munk C.; O'Brien S. J.; Pathak V. K.; Strebel K.; Wain-Hobson S.; Yu X. F.; Yuhki N.; Harris R. S. J. Virol. 2009, 83, 494. |
| [3] | Wedekind J. E. Trends Genet. 2003, 19, 207. |
| [4] | Harris R. S.; Liddament M. T. Nat. Rev. Immunol. 2004, 4, 868. |
| [5] | Kitamura S.; Ode H.; Nakashima M. Nat. Struct. Mol. Biol. 2012, 19, 1005. |
| [6] | Mitra M.; Hercik K.; Byeon I. J. L. Nucleic Acids Res. 2014, 42, 1095. |
| [7] | Chen K. M.; Harjes E.; Gross P. J.; Fahmy A.; Lu Y.; Shindo K. Seibutsu Butsuri 2008, 48, 116 |
| [8] | Burns M. B.; Lackey L.; Carpenter M. A.; Rathore A.; Land A. M.; Leonard B. Nature 2013, 494, 366. |
| [9] | Shi K.; Carpenter M. A.; Kurahashi K.; Harris R. S.; Aihara H. J. Biol. Chem. 2015, 290, 28120. |
| [10] | Xiao X.; Yang H.; Arutiunian V.; Fang Y.; Chen X. S. Nucleic Acids Res. 2017, 45, 7494. |
| [11] | Nathans R.; Cao H.; Sharova N.; Ali A.; Sharkey M.; Stranska R. Nat. Biotechnol. 2008, 26, 1187. |
| [12] | Dang Y.; Wang X.; Esselman W. J.; Zheng Y. H. J. Virol. 2006, 80, 10522. |
| [13] | Stanley B. J.; Ehrlich E. S.; Short L.; Yu Y.; Xiong Y. J. Virol. 2008, 82, 8656. |
| [14] | Seplyarskiy V. B.; Andrianova M. A.; Bazykin G. A. Genome Res. 2017, 27, 175. |
| [15] | Zheng Y. H.; Irwin D.; Kurosu T. J. Virol. 2004, 78, 6073. |
| [16] | Byeon I. J. L.; Ahn J.; Mitra M.; Byeon C. H.; Hercík K.; Hritz J. Nat. Commun. 2013, 4, 1890. |
| [17] | Chelico L.; Prochnow C.; Erie D. A. J. Biol. Chem. 2010, 283, 16195 |
| [18] | Guo Y.; Dong L.; Qiu X.; Wang Y.; Zhang B.; Liu H. Nature 2014, 505 7482 229. |
| [19] | Bohn M. F. Structure 2013, 21, 1042. |
| [20] | Siu K. K.; Sultana A.; Azimi F. Nat. Commun. 2013, 4, 2593. |
| [21] | Matsui M.; Shindo K.; Izumi T.; Io K.; Shinohara M.; Komano J.; Kobayashi M.; Kadowaki N.; Harris R. S.; Takaori-Kondo A. Virol. J. 2014, 11, 122. |
| [22] | Kouno T.; Luengas E. M.; Shigematsu M. Nat. Struct. Mol. Biol. 2015, 22 6 485. |
| [23] | Klarmann G. J. J. Biol. Chem. 2003, 278, 7902. |
| [24] | Furukawa A.; Nagata T.; Matsugami A. Nucleic Acids Symp. Ser. 2009, 53, 87. |
| [25] | Mangeat B.; Turelli P.; Caron G.; Friedli M.; Perrin L.; Trono D. Nature 2003, 4244, 99 |
| [26] | Peng G. J. Exp. Med. 2006, 203, 41. |
| [27] | Chelico L.; Pham P.; Calabrese P.; Goodman M. F. Nat. Struct. Mol. Biol. 2006, 13, 392. |
| [28] | Harjes E.; Gross P. J.; Chen K. M.; Lu Y.; Shindo K. J. Mol. Biol. 2009, 389, 819. |
| [29] | Wichroski M. J. K. Ichiyama; T. M. Rana. J. Biol. Chem. 2005, 280, 8387 |
| [30] | Sheehy A. M.; Gaddis N. C.; Choi J. D.; Malim M. H. Nature 2002, 418, 646. |
| [31] | Bennett R. P.; Presnyak V.; Wedekind J. E.; Smith H. C. J. Biol. Chem. 2008, 283, 7320. |
| [32] | Lu X.; Zhang T. L.; Xu Z. J. Biol. Chem. 2015, 290, 4010. |
| [33] | Dang Y.; Siew L. M.; Wang X. J.; Han Y. X.; Lampen R.; Zheng Y. H. J. Biol. Chem. 2008, 283, 11606. |
| [34] | Jarmuz A.; Chester A.; Bayliss J.; Gisbourne J.; Dunham I.; Scott J. Genomics 2002, 79, 285. |
| [35] | Holden L. G.; Prochnow C.; Chang Y. P. Nature 2008, 456, 121. |
| [36] | Olson M. E.; Li M.; Harris R. S. Chem. Med. Chem. 2013, 8, 112. |
| [37] | Jager S.; Kim D. Y.; Hultquist J. F.; Shindo K.; Larue R. S.; Kwon E. Nature 2012, 481, 371. |
| [38] | Mehle A.; Strack B.; Ancuta P.; Zhang C.; Mcpike M.; Gabuzda D. J. Biol. Chem. 2004, 279, 7792. |
| [39] | Marcsisin S. R.; Engen J. R. J. Mol. Biol. 2010, 402, 892. |
| [40] | Bergeron J. R.; Huthoff H.; Veselkov D. A.; Beavil R. L.; Sanderson M. R. PLoS Pathog. 2010, 6, e1000925. |
| [41] | Reingewertz T. H.; Shalev D. E.; Friedler A. Protein Pept. Lett. 2010, 17, 988. |
| [42] | Liddament M. T.; Brown W. L.; Schumacher A. J.; Harris R. S. Curr. Biol. 2004, 14, 1385. |
| [43] | Lu Z.; Bergeron J. R. C.; AtkinsLu Z.; Bergeron J. R. C.; Atkinson R. A.; Schaller T.; Veselkov D. A.; Oregioni A. Open. Biol. 2013, 3, 130100. |
| [44] | Luo K.; Xiao Z.; Ehrlich E.; Yu Y.; Liu B.; Zheng S. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 11444. |
| [45] | Stenglein M. D.; Burns M. B.; Li M.; Lengyel J.; Harris R. S. Nat. Struct. Mol. Biol. 2010, 17, 222. |
| [46] | Chelico L.; Pham P.; Calabrese P.; Goodman M. F. Nat. Struct. Mol. Biol. 2006, 13, 392. |
| [47] | Ronsard L.; Raja R.; Panwar V.; Saini S.; Mohankumar K.; Sridharan S. Sci. Rep. 2015, 5, 15438. |
| [48] | Yamanaka S.; Balestra M. E.; Ferrell L. D. Proc. Nat. Acad. Sci. U. S. A. 1995, 92, 8483. |
| [49] | Navarro F.; Bollman B.; Chen H. Virology 2005, 333, 374. |
| [50] | Kouno T.; Silvas T. V.; Hilbert B. J.; Shandilya S. M. D.; Bohn M. F.; Kelch B. A.; Royer W. E.; Somasundaran M.; Kurt Yilmaz N.; Matsuo H.; Schiffer C. A. Nat. Commun. 2017, 8, 15024. |
| [51] | Shi K.; Carpenter M. A.; Banerjee S.; Shaban N. M.; Kurahashi K.; Salamango D. J.; McCann J. L.; Starrett G. J.; Duffy J. V.; Demir O.; Amaro R. E.; Harki D. A.; Harris R. S.; Aihara H. Nat. Struct Mol. Biol. 2017, 24, 131. |
| [52] | Fang Y.; Xiao X.; Li S.; Wolfe A.; Chen X. S. J. Mol. Biol. 2018, 430, 87. |
| [53] | Cheng C.; Zhang T.; Wang C. X.; Lan W.; Ding J.; Cao C. Y. Chin. J. Chem. 2018, 36, 1241. |
| [54] | Xiao X.; Li S. X.; Yang H.; Chen X. S. Nat. Commun. 2016, 7, 12193. |
| [55] | Maiti A.; Myint W.; Kanai T.; Delviks-Frankenberry K.; Sierra Rodriguez C.; Pathak V. K.; Schiffer C. A.; Matsuo H. Nat. Commun. 2018, 9, 2460. |
| [56] | Yan X. X.; Lan W. X.; Wang C. X.; Cao C. Y. Chem. Asian J. 2019, 14, 2235. |
| [57] | Bohn J. A.; Thummar K.; York A.; Raymond A.; Brown W. C.; Bieniasz P. D.; Hatziioannou T.; Smith J. L. Nat. Commun. 2017, 8, 1021. |
| [58] | Nadine M. S.; Shi K.; Lauer K. V.; Brown W. L.; Aihara H.; Harris R. S. Mol. Cell 2018, 69, 75. |
| [59] | Matsuoka T.; Nagae T.; Ode H.; Awazu H.; Kurosawa T.; Hamano A.; Matsuoka K.; Hachiya A.; Imahashi M.; Yokomaku Y.; Watanabe N.; Iwatani Y. Nucleic Acids Res. 2018, 46, 10368 |
/
| 〈 |
|
〉 |