

Glycoprotein Identification using Cleavable Bifunctional Probes★
Received date: 2023-06-01
Online published: 2023-08-15
Supported by
National Key R&D Program of China(2021YFA1302603); National Key R&D Program of China(2020YFE0202200); National Natural Science Foundation of China(22125403); National Natural Science Foundation of China(92253304); National Natural Science Foundation of China(32201218); Shenzhen Innovation of Science and Technology Commission(JCYJ20200109141212325); Shenzhen Innovation of Science and Technology Commission(JCYJ20210324120210029); Shenzhen Innovation of Science and Technology Commission(JCYJ20200109140814408); Guangdong Provincial Fund for Distinguished Young Scholars(2019B151502050)
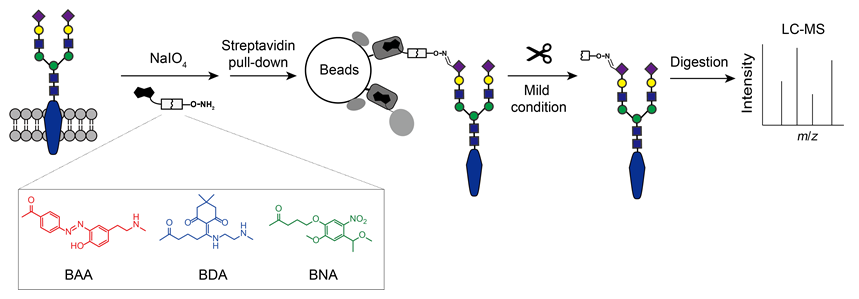
Glycoproteins play important roles in cellular activities and disease development. However, the abundance of glycoproteins is often low in complex biological samples. Thus, it is critical to enrich glycoproteins to achieve highly sensitive mass spectrometry (MS) analysis result. Many methods have been developed to enrich glycoproteins, among which hydrazide chemistry and oxime click chemistry have received increasing attention because of their universality and unbiasedness for labelling glycoproteins. Glycoproteins are captured by covalent binding or high affinity biotin binding on beads in these methods. Therefore, it is difficult to release glycoproteins from beads. Conventional way such as on-bead digestion is too harsh that can co-elute non-specifically bound proteins, endogenously biotinylated proteins and streptavidin on beads. These contaminant proteins would cause background interference in the subsequent glycoprotein identification by MS. In this study, three kinds of cleavable bifunctional probes named Biotin-Azo-Aminooxy (BAA), Biotin-Dde-Aminooxy (BDA) and Biotin-Nbz-Aminooxy (BNA) have been designed and synthesized. The cleavable bifunctional probes allow the release of glycoproteins from beads under mild condition. The mild condition can separate labelled glycoproteins from contaminant proteins to exclude the interference of non-glycoproteins. We evaluated the labelling and enrichment conditions, cleavage efficiency, glycoprotein recovery of these probes. The result showed that the properties of BDA and BNA are excellent. Lastly, BNA was selected to analyze cell surface glycoproteins by MS. Compared with the traditional method of on-bead digestion, the number of non-glycoproteins in this method decreased from 3564 to 2139 by 40.0% and the total Label-free quantitative (LFQ) intensity of glycoproteins increased by 104.2%. Furthermore, the endogenously biotinylated proteins were greatly reduced in cleavage method. The result shows that cleavable bifunctional probes can significantly improve the sensitivity and selectivity of the cell surface glycoprotein identification by MS, which provide tools for deep profiling of glycoproteins in the field of biology and medicine.
Key words: glycoprotein; mass spectrometry; enrichment; oxime click chemistry; cleavable probes
Chang Li , Zhendong Zheng , Jiangnan Zheng , Ruijun Tian . Glycoprotein Identification using Cleavable Bifunctional Probes★[J]. Acta Chimica Sinica, 2023 , 81(12) : 1673 -1680 . DOI: 10.6023/A23050263
| [1] | Pirro, M.; Schoof, E.; van Vliet, S. J.; Rombouts, Y.; Stella, A.; de Ru, A.; Mohammed, Y.; Wuhrer, M.; van Veelen, P. A.; Hensbergen, P. J. J. Proteome Res. 2019, 18, 1125. |
| [2] | Kailemia, M. J.; Xu, G.; Wong, M.; Li, Q.; Goonatilleke, E.; Leon, F.; Lebrilla, C. B. Anal. Chem. 2018, 90, 208. |
| [3] | Venkatakrishnan, V.; Packer, N. H.; Thaysen-Andersen, M. Expert Rev. Respir. Med. 2013, 7, 553. |
| [4] | Shi, Y.; Gao, W.; Lytle, N. K.; Huang, P.; Yuan, X.; Dann, A. M.; Ridinger-Saison, M.; DelGiorno, K. E.; Antal, C. E.; Liang, G.; Atkins, A. R.; Erikson, G.; Sun, H.; Meisenhelder, J.; Terenziani, E.; Woo, G.; Fang, L.; Santisakultarm, T. P.; Manor, U.; Xu, R.; Becerra, C. R.; Borazanci, E.; Von Hoff, D. D.; Grandgenett, P. M.; Hollingsworth, M. A.; Leblanc, M.; Umetsu, S. E.; Collisson, E. A.; Scadeng, M.; Lowy, A. M.; Donahue, T. R.; Reya, T.; Downes, M.; Evans, R. M.; Wahl, G..M.; Pawson, T.; Tian, R.; Hunter, T. Nature 2019, 569, 131. |
| [5] | Campos-da-Paz, M.; Dòrea, J. G.; Galdino, A. S.; Lacava, Z. G. M.; de Fatima Menezes Almeida Santos, M. Recent Pat. Biotechnol. 2018, 12, 269. |
| [6] | Saavedra, D.; Crombet, T. Front Immunol. 2017, 8, 269. |
| [7] | Riley, N. M.; Bertozzi, C. R.; Pitteri, S. J. Mol. Cell Proteomics 2021, 20, 100029. |
| [8] | Suttapitugsakul, S.; Sun, F.; Wu, R. Anal. Chem. 2020, 92, 267. |
| [9] | Zhang, L.; Du, X.; Zeng, Y. Acta Chim. Sinica 2016, 74, 149 (in Chinese). |
| [9] | (张丽霞, 杜秀芳, 曾盈, 化学学报, 2016, 74, 149.) |
| [10] | Li, Y.; Peng, Y.; Lu, H. Acta Chim. Sinica 2021, 79, 705 (in Chinese). |
| [10] | (李月悦, 彭叶, 陆豪杰, 化学学报, 2021, 79, 705.) |
| [11] | Bao, H.; Xie, L.; Lu, H. Se Pu 2016, 34, 1145 (in Chinese). |
| [11] | (包慧敏, 谢力琦, 陆豪杰, 色谱, 2016, 34, 1145.) |
| [12] | Liu, L.; Qin, H.; Ye, M. Se Pu 2021, 39, 1045 (in Chinese). |
| [12] | (刘璐瑶, 秦洪强, 叶明亮, 色谱, 2021, 39, 1045.) |
| [13] | Xiong, Y.; Cheng, M.; Lu, H. Fenxi Huaxue 2021, 49, 1597 (in Chinese). |
| [13] | (熊莹莹, 程孟霞, 陆豪杰, 分析化学, 2021, 49, 1597) |
| [14] | Duan, L.; Zangiabadi, M.; Zhao, Y. Chem. Commun. 2020, 56, 10199. |
| [15] | Shu, Q.; Li, M.; Shu, L.; An, Z.; Wang, J.; Lv, H.; Yang, M.; Cai, T.; Hu, T.; Fu, Y.; Yang, F. Mol. Cell Proteomics. 2020, 19, 672. |
| [16] | Xiao, H.; Chen, W.; Smeekens, J. M.; Wu, R. Nat. Commun. 2018, 9, 1692. |
| [17] | Zeng, Y.; Ramya, T. N.; Dirksen, A.; Dawson, P. E.; Paulson, J. C. Nat. Methods. 2009, 6, 207. |
| [18] | Zhang, Y.; Yu, M.; Zhang, C.; Ma, W.; Zhang, Y.; Wang, C.; Lu, H. Anal. Chem. 2014, 86, 7920. |
| [19] | Szychowski, J.; Mahdavi, A.; Hodas, J. J.; Bagert, J. D.; Ngo, J. T.; Landgraf, P.; Dieterich, D. C.; Schuman, E. M.; Tirrell, D. A. J. Am. Chem. Soc. 2010, 132, 18351. |
| [20] | Yang, Y.; Hahne, H.; Kuster, B.; Verhelst, S. H. Mol. Cell Proteomics 2013, 12, 237. |
| [21] | Beard, H. A.; Korovesis, D.; Chen, S.; Verhelst, S. H. L. Mol. Omics 2021, 17, 197. |
| [22] | Eom, T.; Khan, A. Org. Biomol. Chem. 2020, 18, 420. |
| [23] | Mnatsakanyan, R.; Markoutsa, S.; Walbrunn, K.; Roos, A.; Verhelst, S. H. L.; Zahedi, R. P. Nat. Commun. 2019, 10, 2195. |
| [24] | Chang, T. C.; Adak, A. K.; Lin, T. W.; Li, P. J.; Chen, Y. J.; Lai, C. H.; Liang, C. F.; Chen, Y. J.; Lin, C. C. Bioorg. Med. Chem. 2016, 24, 1216. |
| [25] | Chen, W.; Wang, S.; Adhikari, S.; Deng, Z.; Wang, L.; Chen, L.; Ke, M., Yang, P.; Tian, R. Anal. Chem. 2016, 88, 4864. |
| [26] | Gao, W.; Zhang, Q.; Su, Y.; Huang, P.; Lu, X.; Gong, Q.; Chen, W.; Xu, R.; Tian, R. Analyst 2020, 145, 6441. |
| [27] | Pace, C. L.; Muddiman, D. C. J. Am. Soc. Mass Spectrom. 2020, 10, 1021. |
| [28] | Frankenfield, A. M.; Fernandopulle, M. S.; Hasan, S.; Ward, M. E.; Hao, L. Anal. Chem. 2020, 92, 15437. |
| [29] | Chu, B.; He, A.; Tian, Y.; He, W.; Chen, P.; Hu, J.; Xu, R.; Zhou, W.; Zhang, M.; Yang, P.; Li, S. S. C.; Sun, Y.; Li, P.; Hunter, T.; Tian, R. Proc. Natl. Acad. Sci. U. S. A. 2018, 92, E8863. |
/
| 〈 |
|
〉 |