

Chinese Journal of Organic Chemistry >
Solid Phase Synthesis and Property of Signature Motif Ⅲ in Peptide Transporter
Received date: 2017-08-24
Revised date: 2017-10-11
Online published: 2017-10-16
Supported by
Project supported by the National Natural Science Foundation of China (Nos. 21572046, 21172054) and the Foundation of Scientific and Technological Project of Henan Province (No. 162102210197).
The peptide transporter family in human body is critical for the transport of peptides and drugs, and the conservative sequences in the peptide transporters play an important role in maintaining its structure and function. In order to understand the function of consensus peptides in peptide transporter and promote the application of oligopeptides in pharmaceutical and medical fields, the signature motif Ⅲ (FYLSINAGS) and its four mutants were synthesized by Fmoc solid phase synthesis method. The products were identified using mass spectrometry, and purified by RP-HPLC. The interaction between peptide and DNA was detected by UV and fluorescence spectrometry. The experimental and structural simulation results showed that the dominant role of electrostatic interaction and intercalation between oligopeptide FYLSINAGG and DNA was related to the concentration of oligopeptide FYLSINAGG, the interaction of FYGLINAGG containing helix structure and DNA was enhanced for the generation of complex, and the interaction between FYGLINKGG hasing helix structure or FYGLINSGG and DNA was attenuated. These results indicated that the serine residue in C-terminal of FYGSINAGS, and the number and position of serine in peptides had great influence on the structure of oligopeptide and its interaction with DNA. The mutation of serine into hydrophobic amino acids is beneficial to form helical structure of oligopeptides and enhance the embedded intercalation with DNA. Thus, serine residues in signature motif are important functional residues, especially at the C-terminus.
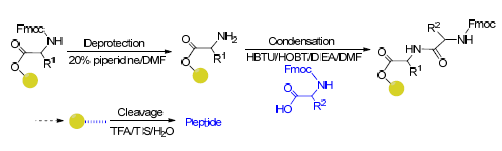
Key words: oligopeptide; solid phase synthesis; interaction; signature motif
Zhao Dongxin, Lü Mingxiu, Ma Li, Lu Kui . Solid Phase Synthesis and Property of Signature Motif Ⅲ in Peptide Transporter[J]. Chinese Journal of Organic Chemistry, 2018 , 38(1) : 266 -271 . DOI: 10.6023/cjoc201708053
[1] (a) Frazer, K. A.; Sheehan, J. B.; Stokowski, R. P.; Chen, X.; Hosseini, R.; Cheng, J. F.; Fodor, S. P.; Cox, D. R.; Patil, N. Genome Res. 2001, 11, 1651.
(b) Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, D.; Kent, W. J.; Mattick, J. S.; Haussler, D. Science 2004, 304, 1321.
(c) Fire, A.; Xu, S.; Montgomery, M. K.; Kostas, S. A.; Driver, S. E.; Mello, C. C. Nature 1998, 391, 806.
(d) Florence, W.; Magdalena, Z. G. Nat. Cell Biol. 2000, 2, 70.
[2] (a) Ocheltree, S. M.; Keep, R. F.; Shen, H.; Yang, D. L.; Hughes, B. A.; Smith D. E. Pharmacol. Res. 2003, 20, 1364.
(b) Zhao, D. X.; Lu, K. Biosci. Trends 2015, 9, 207.
(c) Smith, D. E.; Clémencon, B.; Hediger, M. A. Mol. Aspects Med. 2013, 34, 323.
(d) Charrière, G. M.; Eddie Ip, W. K.; Dejardin, S.; Boyer, L.; Sokolovska, A.; Cappillino, M. P.; Cherayil, B. J.; Podolsky, D. K; Kobayashi, K. S.; Silverman, N.; Lacy-Hulbert, A.; Stuart, L. M. J. Biol. Chem. 2010, 285, 20147.
(e) Fei, Y. J.; Liu, W.; Prasad, P. D.; Kekuda, R.; Oblak, T. G.; Ganapathy, V.; Leibach, F. H. Biochemistry 1997, 36, 452.
[3] (a) Daniel, H.; Kottra, G. Pflug. Arch. Eur. J. Phy. 2004, 447, 610.
(b) Gindullis, F.; Rose, A.; Patel, S.; Meier, I. BMC Genomics 2002, 3, 9.
(c) Li, S. B; Qian, Q.; Fu, Z. M.; Zeng, D. L.; Meng, X. B.; Kyozuka, J.; Maekawa, M.; Zhu, X. D.; Zhang, J.; Li, J. Y.; Wang, Y. H. Plant J. 2009, 58, 592.
[4] (a) Cai, C. Q.; Chen, X. M.; Ge, F. Spectrochim. Acta A 2010, 76, 202.
(b) Zhao, D. X.; Ma, L.; Lu, K. Lett. Org. Chem. 2015, 12, 459.
(c) Wu, J. H.; Wei, L.; Zhao, M.; Wang, Y. J.; Kang, G. F.; Peng, S. Q. Med. Chem. Res. 2012, 21, 116.
(d) Li, N.; Ma, Y.; Yang, C.; Guo, L. P.; Yang, X. R. Biophys. Chem. 2005, 116, 199.
(e) Iwasaki, Y.; Kimura, M.; Yamada, A.; Mutoh, Y.; Tateishi, M.; Arii, H. Inorg. Chem. Commun. 2011, 14, 1461.
[5] (a) Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. J. Chem. Theory Comput. 2014, 10, 4745.
(b) Maupetit, J.; Derreumaux, P.; Tufféry, P. Nucleic Acids Res. 2009, 37, 498.
[6] (a) Long, E. C.; Barton, J. K. Acc. Chem. Res. 1990, 23, 271.
(b) Zhou, C. Y.; Zhao, Y.; Wu, Y. B.; Yin, C. X.; Yang, P. J. Inorg. Biochem. 2007, 101, 10.
(c) Guo, Q.; Li, L. Z.; Dong, J. F.; Liu, H. Y.; Xue, Z. C.; Xv, T. Acta Chim. Sinica 2012, 70, 1617(in Chinese).(郭琼, 李连之, 董建方, 刘鸿雁, 薛泽春, 许涛, 化学学报, 2012, 70, 1617.)
[7] (a) Gao, E. J.; Zhu, M. C.; Yin, H. X.; Liu, L.; Wu, Q.; Sun, Y. G. J. Inorg. Biochem. 2008, 102, 1958.
(b) Zhao, D. X.; Ma, L.; Lu, K.; Wu, J. Z.; He, J. Res. Chem. Intermediat 2015, 41, 8591.
(c) Chen, D. D.; Wu, Q.; Wang, J.; Wang, Q.; Qiao, H. Spectrochim. Acta A 2015, 135, 511.
(d) Guo, Y.; Liiu, B. S.; Li, Z. Y.; Zhang, L. H.; Lv, Y. K. Chin. J. Lumin. 2014, 35, 1114(in Chinese).(郭莹, 刘保生, 李志云, 张丽惠, 吕运开, 发光学报, 2014, 35, 1114.)
(e) Li, H.; Le, X. Y.; Wu, J. Z.; Liu, J.; Ji, L. N.; Jiang, X.; Li, W. S.; Xu, Z. H. Acta Chim. Sinica 2003, 61, 245.
[8] (a) Geng, S. G.; Cui, Y. R.; Liu, Q. F. J. Lumin. 2013, 141, 144.
(b) Zhao, D. X.; Lu, K. Heterocycles 2016, 92, 252.
(c) Muro, P. D.; Beltramini, M.; Nikolov, P.; Retkova, I.; Salvato, B.; Ricchelli, F. Z. Naturforsch. B 2002, 57, 1084.
[9] (a) Zhao, D. X; Lu, K. J. Mol. Model. 2015, 21, 299.
(b) Pires, D. A. T.; Bemquerer, M. P.; Nascimento, C. J. D. Int. J. Pept. Res. Ther. 2014, 20, 53.
(c) Amblard, M.; Fehrentz, J. A.; Martinez, J.; Subra, G. Mol. Biotechnol. 2006, 33, 239.
/
| 〈 |
|
〉 |