化学学报 ›› 2022, Vol. 80 ›› Issue (6): 714-723.DOI: 10.6023/A22010017 上一篇 下一篇
研究论文
刘雨泽a, 李昆华a, 黄佳兴a, 于曦a,b,*( ), 胡文平a,b,*(
), 胡文平a,b,*( )
)
投稿日期:2022-01-10
发布日期:2022-07-07
通讯作者:
于曦, 胡文平
基金资助:
Yuze Liua, Kunhua Lia, Jiaxing Huanga, Xi Yua,b( ), Wenping Hua,b(
), Wenping Hua,b( )
)
Received:2022-01-10
Published:2022-07-07
Contact:
Xi Yu, Wenping Hu
Supported by:文章分享

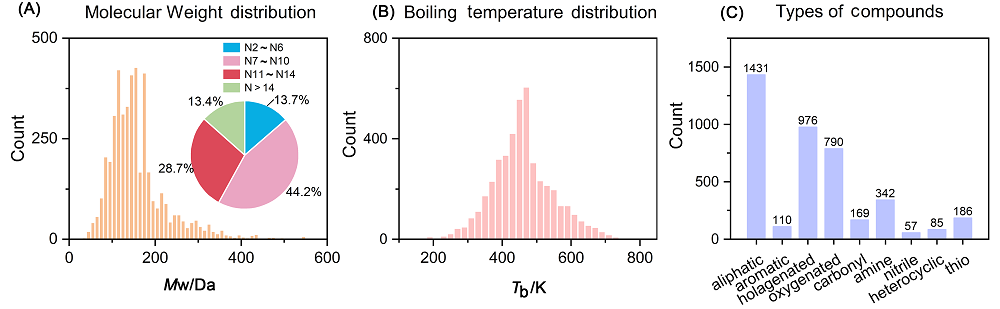
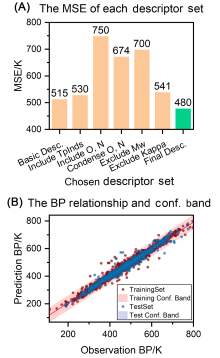
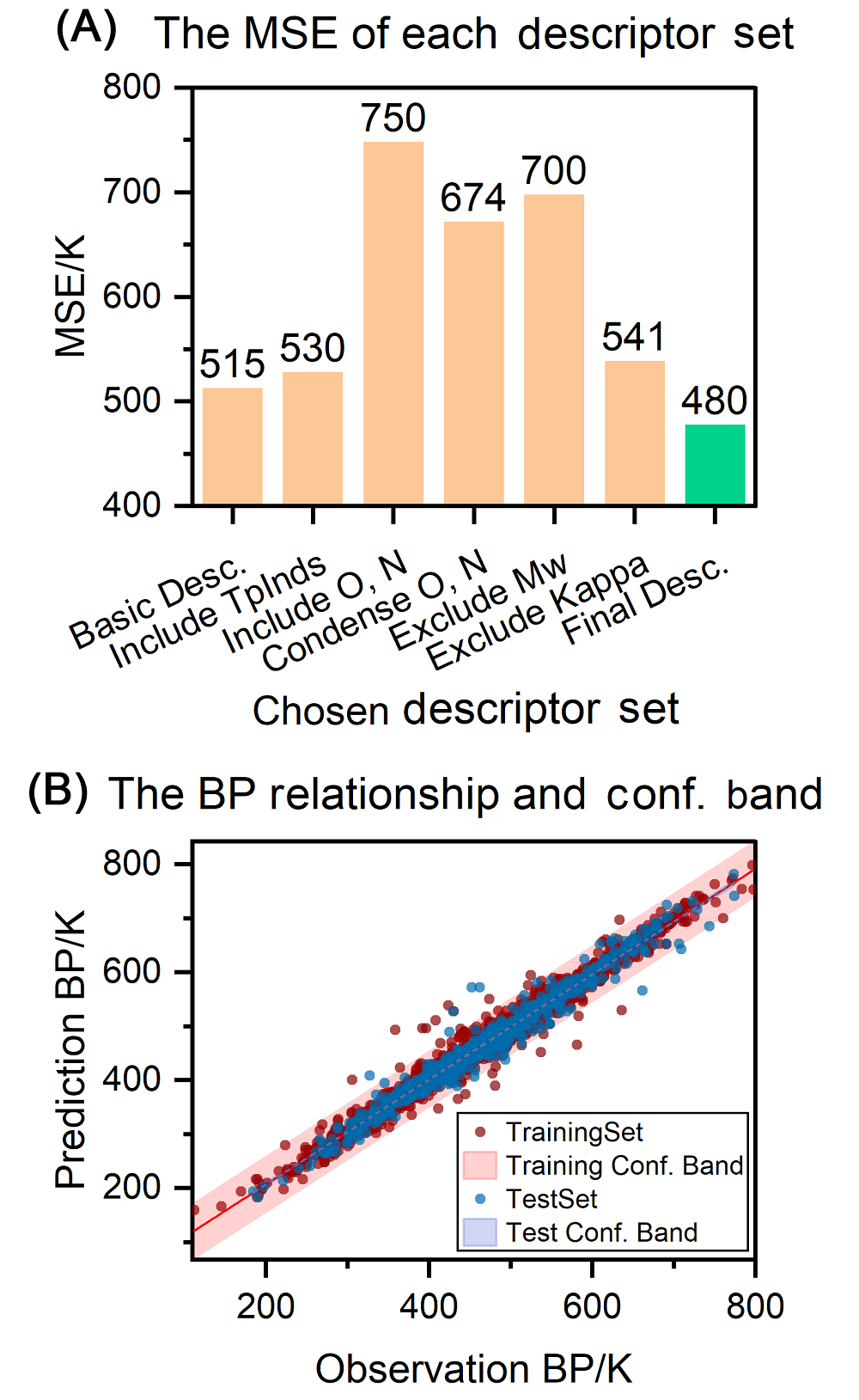
沸点(BP)是有机分子液体的基本物理化学量, 也是化学工业生产中的重要参数. 有机分子的沸点由分子结构决定, 呈现复杂的结构-沸点关系, 函数法(Function Method)、基团贡献法(Group Contribution Method)等传统方法无法应对复杂多样有机分子结构的预测, 应用范围狭窄, 预测精度低. 本研究中, 我们利用基于人工神经网络(ANN)和支持向量机(SVM)的多组件学习器实现有机分子沸点的精准预测. 我们构建了基于可解释性描述符的ANN、基于相关性描述符的ANN及基于复合分子指纹的SVM三个异质模型, 并通过包含4550个各种类别的有机分子沸点的数据集进行训练得到了三个异质性学习器, 最后集成三个学习器对有机分子沸点进行预测. 相比于传统方法和此前的定量结构性质关系(QSPR)模型, 多组件模型结合了三种模型的优点, 展现出很好的预测精度和泛化能力以及低的过拟合, 实现了对多种类型有机分子的沸点的有效预测.
刘雨泽, 李昆华, 黄佳兴, 于曦, 胡文平. 多组件学习器实现有机分子沸点的精准预测[J]. 化学学报, 2022, 80(6): 714-723.
Yuze Liu, Kunhua Li, Jiaxing Huang, Xi Yu, Wenping Hu. Accurate Prediction of the Boiling Point of Organic Molecules by Multi-Component Heterogeneous Learning Model[J]. Acta Chimica Sinica, 2022, 80(6): 714-723.

| No. | Name | Type | Detail | |
|---|---|---|---|---|
| 1 | Log P | Molecular properties | Octanol-water partition coeff. | |
| 2 | nROH | Functional group | Number of hydroxyl groups | |
| 3 | nROR | Functional group | Number of ethers | |
| 4 | nR=O | Functional group | Number of carbonyl | |
| 5 | nRNH2 | Functional group | Number of 1st amine | |
| 6 | nRNHR | Functional group | Number of 2st amine | |
| 7 | nRNR2 | Functional Group | Number of 3st amine | |
| 8 | nRCN | Functional Group | Number of nitriles | |
| 9 | nSatom | Constitutional indices | Number of Surfur atom | |
| 10 | nFatom | Constitutional indices | Number of Fluorine atom | |
| 11 | nClatom | Constitutional indices | Number of Chlorine atom | |
| 12 | nBratom | Constitutional indices | Number of Bromine atom | |
| 13 | nIatom | Constitutional indices | Number of Iodine atom | |
| 14 | nCatom | Constitutional indices | Number of carbon atom | |
| 15 | HA | Structural Parameter | Number of acceptor atom for H-bond | |
| 16 | HD | Structural Parameter | Number of donor atom for H-bond | |
| 17 | nCharge | Constitutional indices | Total charge | |
| 18 | Chi2n | Topological indices | 2-Path kier shape index | |
| 19 | AromR | Ring descriptors | Aromatic Ratio |
| No. | Name | Type | Detail | |
|---|---|---|---|---|
| 1 | Log P | Molecular properties | Octanol-water partition coeff. | |
| 2 | nROH | Functional group | Number of hydroxyl groups | |
| 3 | nROR | Functional group | Number of ethers | |
| 4 | nR=O | Functional group | Number of carbonyl | |
| 5 | nRNH2 | Functional group | Number of 1st amine | |
| 6 | nRNHR | Functional group | Number of 2st amine | |
| 7 | nRNR2 | Functional Group | Number of 3st amine | |
| 8 | nRCN | Functional Group | Number of nitriles | |
| 9 | nSatom | Constitutional indices | Number of Surfur atom | |
| 10 | nFatom | Constitutional indices | Number of Fluorine atom | |
| 11 | nClatom | Constitutional indices | Number of Chlorine atom | |
| 12 | nBratom | Constitutional indices | Number of Bromine atom | |
| 13 | nIatom | Constitutional indices | Number of Iodine atom | |
| 14 | nCatom | Constitutional indices | Number of carbon atom | |
| 15 | HA | Structural Parameter | Number of acceptor atom for H-bond | |
| 16 | HD | Structural Parameter | Number of donor atom for H-bond | |
| 17 | nCharge | Constitutional indices | Total charge | |
| 18 | Chi2n | Topological indices | 2-Path kier shape index | |
| 19 | AromR | Ring descriptors | Aromatic Ratio |
| No. | Descriptora | Typeb | Detail | |
|---|---|---|---|---|
| 1 | Sv | Constitutional indices | Sum of VDW volumes | |
| 2 | Sp | Constitutional indices | Sum of polarizabilities | |
| 3 | nBO | Constitutional indices | Number of non-H bonds | |
| 4 | SCBO | Constitutional indices | Sum of conventional bond orders | |
| 5 | nH | Constitutional indices | Numbers of Hydrogen atoms | |
| 6 | pilD | Walk and path counts | Conventional bond order ID number | |
| 7 | IDET | Information indices | Total information content on the distance equality | |
| 8 | HVcpx | Information indices | graph vertex complexity index | |
| 9 | TlC1 | Information indices | Total Information Content index | |
| 10 | ClC0 | Information indices | Complementary Information Content index | |
| 11 | ATS4m | 2D autocorrelations | B-M autocorrelation of lag 4 (log function) weighted by mass | |
| 12 | ATS2p | 2D autocorrelations | B-M autocorrelation of lag 2 (log function) weighted by polarizability | |
| 13 | ATS1li | 2D autocorrelations | B-M autocorrelation of lag 1 (log function) weighted by ionization potential | |
| 14 | ATS3i | 2D autocorrelations | B-M autocorrelation of lag 3 (log function) weighted by ionization potential | |
| 15 | ATSC1i | 2D autocorrelations | Centred B-M autocorrelation of lag 1 weighted by ionization potential | |
| 16 | MATS1i | 2D autocorrelations | Moran autocorrelation of lag 1 weighted by ionization potential | |
| 17 | GGl10 | 2D autocorrelations | topological charge index of order 10 | |
| 18 | SpMin3_Bh(m) | Burden eigenvalues | smallest eigenvalue n. 3 of Burden matrix weighted by mass | |
| 19 | SpMin4_Bh(m) | Burden eigenvalues | smallest eigenvalue n. 4 of Burden matrix weighted by mass | |
| 20 | P_VSA_m_2 | P_VSA-like descriptors | P_VSA-like on mass, bin 2 | |
| 21 | P_VSA_e_2 | P_VSA-like descriptors | P_VSA-like on Sanderson electronegativity, bin 2 | |
| 22 | P_VSA_p_1 | P_VSA-like descriptors | P_VSA-like on polarizability, bin 1 | |
| 23 | P_VSA_ppp_L | P_VSA-like descriptors | P_VSA-like on potential pharmacophore points, L-lipophilic | |
| 24 | P_VSA_ppp_ar | P_VSA-like descriptors | P_VSA-like on potential pharmacophore points, ar-aromatic atoms | |
| 25 | P_VSA_charge_6 | P_VSA-like descriptors | P_VSA-like on partial charges, bin 6 | |
| 26 | nCbH | Functional group counts | number of unsubstituted benzene C(sp2) | |
| 27 | CATS2D_03_LL | Pharmacophore descriptors | CATS2D Lipophilic-Lipophilic at lag 03 | |
| 28 | Qpos | Charge descriptors | total positive charge | |
| 29 | RPCG | Charge descriptors | relative positive charge | |
| 30 | MLOGP2 | Molecular properties | squared logP | |
| 31 | LOGPcons | Molecular properties | Octanol-water partition coeff. (consensus) | |
| 32 | PDI | Molecular properties | packing density index | |
| 33 | BLTD48 | Molecular properties | Verhaar Daphnia base-line toxicity from MLOGP (mmol/L) | |
| 34 | DLS_cons | Drug-like indices | DRAGON consensus drug-like score | |
| 35 | MDEC-23 | MDE descriptors | molecular distance edge between all secondary and tertiary carbons |
| No. | Descriptora | Typeb | Detail | |
|---|---|---|---|---|
| 1 | Sv | Constitutional indices | Sum of VDW volumes | |
| 2 | Sp | Constitutional indices | Sum of polarizabilities | |
| 3 | nBO | Constitutional indices | Number of non-H bonds | |
| 4 | SCBO | Constitutional indices | Sum of conventional bond orders | |
| 5 | nH | Constitutional indices | Numbers of Hydrogen atoms | |
| 6 | pilD | Walk and path counts | Conventional bond order ID number | |
| 7 | IDET | Information indices | Total information content on the distance equality | |
| 8 | HVcpx | Information indices | graph vertex complexity index | |
| 9 | TlC1 | Information indices | Total Information Content index | |
| 10 | ClC0 | Information indices | Complementary Information Content index | |
| 11 | ATS4m | 2D autocorrelations | B-M autocorrelation of lag 4 (log function) weighted by mass | |
| 12 | ATS2p | 2D autocorrelations | B-M autocorrelation of lag 2 (log function) weighted by polarizability | |
| 13 | ATS1li | 2D autocorrelations | B-M autocorrelation of lag 1 (log function) weighted by ionization potential | |
| 14 | ATS3i | 2D autocorrelations | B-M autocorrelation of lag 3 (log function) weighted by ionization potential | |
| 15 | ATSC1i | 2D autocorrelations | Centred B-M autocorrelation of lag 1 weighted by ionization potential | |
| 16 | MATS1i | 2D autocorrelations | Moran autocorrelation of lag 1 weighted by ionization potential | |
| 17 | GGl10 | 2D autocorrelations | topological charge index of order 10 | |
| 18 | SpMin3_Bh(m) | Burden eigenvalues | smallest eigenvalue n. 3 of Burden matrix weighted by mass | |
| 19 | SpMin4_Bh(m) | Burden eigenvalues | smallest eigenvalue n. 4 of Burden matrix weighted by mass | |
| 20 | P_VSA_m_2 | P_VSA-like descriptors | P_VSA-like on mass, bin 2 | |
| 21 | P_VSA_e_2 | P_VSA-like descriptors | P_VSA-like on Sanderson electronegativity, bin 2 | |
| 22 | P_VSA_p_1 | P_VSA-like descriptors | P_VSA-like on polarizability, bin 1 | |
| 23 | P_VSA_ppp_L | P_VSA-like descriptors | P_VSA-like on potential pharmacophore points, L-lipophilic | |
| 24 | P_VSA_ppp_ar | P_VSA-like descriptors | P_VSA-like on potential pharmacophore points, ar-aromatic atoms | |
| 25 | P_VSA_charge_6 | P_VSA-like descriptors | P_VSA-like on partial charges, bin 6 | |
| 26 | nCbH | Functional group counts | number of unsubstituted benzene C(sp2) | |
| 27 | CATS2D_03_LL | Pharmacophore descriptors | CATS2D Lipophilic-Lipophilic at lag 03 | |
| 28 | Qpos | Charge descriptors | total positive charge | |
| 29 | RPCG | Charge descriptors | relative positive charge | |
| 30 | MLOGP2 | Molecular properties | squared logP | |
| 31 | LOGPcons | Molecular properties | Octanol-water partition coeff. (consensus) | |
| 32 | PDI | Molecular properties | packing density index | |
| 33 | BLTD48 | Molecular properties | Verhaar Daphnia base-line toxicity from MLOGP (mmol/L) | |
| 34 | DLS_cons | Drug-like indices | DRAGON consensus drug-like score | |
| 35 | MDEC-23 | MDE descriptors | molecular distance edge between all secondary and tertiary carbons |

| Fingerprint | Length | Radical | MSE |
|---|---|---|---|
| ECFPa | 1024 | 2-2 | 378 |
| ECFP | 2048 | 2-2 | 398 |
| ECFP | 2048 | 2-4 | 420 |
| PFPb | 1024 | 2-6 | 685 |
| Fingerprint | Length | Radical | MSE |
|---|---|---|---|
| ECFPa | 1024 | 2-2 | 378 |
| ECFP | 2048 | 2-2 | 398 |
| ECFP | 2048 | 2-4 | 420 |
| PFPb | 1024 | 2-6 | 685 |
| Model composition | R2train | R2test | MAEtrain | MAEtest | MSEtrain | MSEtest |
|---|---|---|---|---|---|---|
| Model A | 0.9885 | 0.9793 | 9.23 | 10.84 | 186.7 | 333.9 |
| Model B | 0.995 | 0.9833 | 5.61 | 8.53 | 81.4 | 270.1 |
| Model C | 0.9951 | 0.9789 | 6.01 | 10.45 | 83.2 | 350.1 |
| Heterogenousa | 0.9953 | 0.9872 | 5.52 | 7.95 | 77.8 | 209.8 |
| Heterovotedb | 0.9953 | 0.9864 | 5.55 | 8.1 | 73.8 | 214.6 |
| Homogenousc | 0.9955 | 0.9848 | 5.16 | 7.96 | 79.9 | 245.6 |
| Model composition | R2train | R2test | MAEtrain | MAEtest | MSEtrain | MSEtest |
|---|---|---|---|---|---|---|
| Model A | 0.9885 | 0.9793 | 9.23 | 10.84 | 186.7 | 333.9 |
| Model B | 0.995 | 0.9833 | 5.61 | 8.53 | 81.4 | 270.1 |
| Model C | 0.9951 | 0.9789 | 6.01 | 10.45 | 83.2 | 350.1 |
| Heterogenousa | 0.9953 | 0.9872 | 5.52 | 7.95 | 77.8 | 209.8 |
| Heterovotedb | 0.9953 | 0.9864 | 5.55 | 8.1 | 73.8 | 214.6 |
| Homogenousc | 0.9955 | 0.9848 | 5.16 | 7.96 | 79.9 | 245.6 |


| Type of compound | N | Descriptor | Method | R2 | S | Reference |
|---|---|---|---|---|---|---|
| Alkanes | 94 | Tls(W, P) | MLR | 0.97 | [ | |
| Alkanes | 74 | Tls(5) | MLR | 0.999 | 1.86 | [ |
| Alkanes(C2~C7) | 72 | Tls(LOVI’s) | RA | 0.994 | 3.9 | [ |
| Furans, thiophenestrahydrofuran | 209 | Tls, electronic, geometrical | RA | 0.969 | 11.2 | [ |
| Alkanes, alcohols | 245 | Atom-type E | MLR | 8 | [ | |
| Alcohols | 58 | Weighted path related Tls | MRA | 0.978 | 3.64 | [ |
| Diverse organic compounds | 298 | Constitutional, topological, geometrical and CPSA(8) | MLR | 0.954 | 16.15 | [ |
| Hydrocarbons | 143 | Tls | MLR | 0.9821 | 7.2549 | [ |
| Diverse organic compounds (contain N, O, F, Cl, Br and I) | 612 | Constitutional, topological, geometrical and CPSA(8) | MLR | 0.965 | 15.5 | [ |
| Diverse organic compounds | 450 | Molecular weight and specific gravity | RBF | 18.78 | [ | |
| Single ring, fused rings, halogens OH, COOH, COOR, CON, CN, NH2 and NO2 | 155 | Tls | BMLR | 0.9864 | 9.1 | [ |
| Diverse organic compounds | 14216 | Mathematical Selection | ANN | 0.943 | 22 | [ |
| Diverse organic compounds | 4550 | Tls, Constitutional, Fingerprints | Composite ML | 0.9953 | 10.0873 | Present work |
| Type of compound | N | Descriptor | Method | R2 | S | Reference |
|---|---|---|---|---|---|---|
| Alkanes | 94 | Tls(W, P) | MLR | 0.97 | [ | |
| Alkanes | 74 | Tls(5) | MLR | 0.999 | 1.86 | [ |
| Alkanes(C2~C7) | 72 | Tls(LOVI’s) | RA | 0.994 | 3.9 | [ |
| Furans, thiophenestrahydrofuran | 209 | Tls, electronic, geometrical | RA | 0.969 | 11.2 | [ |
| Alkanes, alcohols | 245 | Atom-type E | MLR | 8 | [ | |
| Alcohols | 58 | Weighted path related Tls | MRA | 0.978 | 3.64 | [ |
| Diverse organic compounds | 298 | Constitutional, topological, geometrical and CPSA(8) | MLR | 0.954 | 16.15 | [ |
| Hydrocarbons | 143 | Tls | MLR | 0.9821 | 7.2549 | [ |
| Diverse organic compounds (contain N, O, F, Cl, Br and I) | 612 | Constitutional, topological, geometrical and CPSA(8) | MLR | 0.965 | 15.5 | [ |
| Diverse organic compounds | 450 | Molecular weight and specific gravity | RBF | 18.78 | [ | |
| Single ring, fused rings, halogens OH, COOH, COOR, CON, CN, NH2 and NO2 | 155 | Tls | BMLR | 0.9864 | 9.1 | [ |
| Diverse organic compounds | 14216 | Mathematical Selection | ANN | 0.943 | 22 | [ |
| Diverse organic compounds | 4550 | Tls, Constitutional, Fingerprints | Composite ML | 0.9953 | 10.0873 | Present work |
| [1] |
Walker, J. J. Chem. Soc. 1894, 65, 193.
doi: 10.1039/CT8946500193 |
| [2] |
Joback, K. G.; Reid, R. C. Chem. Eng. Commun. 1987, 57, 230.
|
| [3] |
Wiener, H. J. Am. Chem. Soc. 1947, 69, 17.
doi: 10.1021/ja01193a005 |
| [4] |
Katritzky, A. R.; Kuanar, M.; Slavov, S.; Hall, C. D.; Karelson, M.; Kahn, I.; Dobchev, D. A. Chem. Rev. 2010, 110, 5714.
doi: 10.1021/cr900238d pmid: 20731377 |
| [5] |
Zhu, B.; Wu, R.; Yu, X. Acta Chim. Sinica 2020, 78, 1366. (in Chinese)
doi: 10.6023/A20070306 |
|
(朱博阳, 吴睿龙, 于曦, 化学学报, 2020, 78, 1366.)
doi: 10.6023/A20070306 |
|
| [6] |
Wei, J.; Chu, X.; Sun, X.; Xu, K.; Deng, H.; Chen, J.; Wei, Z.; Lei, M. InfoMat 2019, 1, 338.
doi: 10.1002/inf2.12028 |
| [7] |
Song, Z.; Chen, X.; Meng, F.; Cheng, G.; Wang, C.; Sun, Z.; Yin, W. J. Chin. Phys. B 2020, 29, 68.
|
| [8] |
Wu, W.; Sun, Q. Scientia Sinica: Physica, Mechanica et Astronomica 2018, 48, 58. (in Chinese)
|
|
(吴炜, 孙强, 中国科学: 物理学, 力学, 天文学, 2018, 48, 58.)
|
|
| [9] |
Liu, Y. D.; Yang, Q.; Li, Y.; Zhang, L.; Luo, S. Z. Chin. J. Org. Chem. 2020, 40, 3812. (in Chinese)
doi: 10.6023/cjoc202006051 |
|
(刘伊迪, 杨骐, 李遥, 张龙, 罗三中, 有机化学, 2020, 40, 3812.)
doi: 10.6023/cjoc202006051 |
|
| [10] |
Fissa, M. R. J. Mol. Graph. Model. 2019, 87, 109.
doi: 10.1016/j.jmgm.2018.11.013 |
| [11] |
Goll, E. S.; Jurs, P. C. J. Chem. Inf. Model. 1999, 39, 974.
|
| [12] |
Beck, B.; Breindl, A.; Clark, T. J. Chem. Infor. Comp. Sci. 2000, 40, 1046.
|
| [13] |
Chalk, A. J.; Beck, B.; Clark, T. J. Chem. Infor. Comp. Sci. 2001, 41, 457.
|
| [14] |
Gharagheizi, F.; Mirkhani, S. A.; Ilani-Kashkouli, P.; Mohammadi, A. H.; Ramjugernath, D.; Richon, D. Fluid Phase Equilib. 2013, 354, 250.
doi: 10.1016/j.fluid.2013.06.034 |
| [15] |
Yaws, C. L. Yaws' Critical Property Data for Chemical Engineers and Chemists, 2012, http://app.knovel.com/hotlink/toc/id:kpYCPDCECD/yaws-critical-property/yaws-critical-property
|
| [16] |
PubChem-https://pubchem.ncbi.nlm.nih.gov/
|
| [17] |
Mauri, A. Ecotoxicological QSARs, Methods in Pharmacology and Toxicology, Roy, K., New York, 2020, pp. 801-820.
|
| [18] |
Kubic, W. L.; Jenkins, R. W.; Moore, C. M.; Semelsberger, T. A.; & Sutton, A. D. Ind. Eng. Chem. Res. 2017, 56, 12236.
doi: 10.1021/acs.iecr.7b02753 |
| [19] |
Kier, L. B.; Hall, L. H. In Molecular ConnectiVity in Chemistry and Drug Research, New York, 1976, pp. 27-39.
|
| [20] |
Dash, M.; Liu, H. Intell. Data Anal., 1997, 1, 131.
doi: 10.3233/IDA-1997-1302 |
| [21] |
Roy, P. P.; Leonard, J. T.; Roy, K. Chemometr. Intell. Lab. Syst. 2008, 90, 31.
doi: 10.1016/j.chemolab.2007.07.004 |
| [22] |
MacKay, J. C. Neural Comput. 1992, 4, 415
doi: 10.1162/neco.1992.4.3.415 |
| [23] |
Zhou, Z. H. Ensemble Methods: Foundations and Algorithms, Chapman and Hall/CRC, 2012, p. 129
|
| [24] |
Zhou, L.; Wang, B.; Jiang, J.; Pan, Y.; & Wang, Q. Chemometr. Intell. Lab. Syst. 2017, 167, 190.
doi: 10.1016/j.chemolab.2017.06.009 |
| [25] |
Robert, T. J. R. Stat. Soc. Ser. A 1996, 58, 267.
|
| [26] |
Sheela, K.; Deepa, S. N. Math. Probl. Eng. 2013, 2013, 11.
|
| [27] |
Eriksson, L.; Jaworska, J.; Worth, A. P.; Cronin, M. T. D.; McDowell, R. M.; Gramatica, P. Environ. Health Perspect. 2003, 111, 1361.
doi: 10.1289/ehp.5758 |
| [28] |
Needham, D. E.; Wei, I. C.; Seybold, P. G. J. Am. Chem. Soc. 1988, 110, 4186.
doi: 10.1021/ja00221a015 |
| [29] |
Balaban, A. T.; Ciubotariu, D.; Medeleanu, M. J. Chem. Infor. Comp. Sci. 1991, 313, 517.
|
| [30] |
Stanton, D. T. J. Chem. Infor. Comp. Sci. 2000, 40, 81.
|
| [31] |
Hall, L. H.; Kier, L. B. J. Chem. Infor. Comp. Sci. 1995, 35, 1039.
|
| [32] |
Randić, M.; Balaban, A. T.; Basak, S. J. Chem. Infor. Comp. Sci. 2001, 41, 593.
|
| [33] |
Katritzky, A. R.; Mu, L.; Lobanov, V. S.; Karelson, M. J. Phys. Chem. 1996, 100, 10400.
doi: 10.1021/jp953224q |
| [34] |
Zhou, C. Y.; Nie, C. M.; Li, S.; Li, Z. H. J. Comput. Chem. 2007, 28, 2413.
doi: 10.1002/jcc.20540 |
| [35] |
Katritzky, A. R.; Lobanov, V. S.; Karelson, M. J. Chem. Infor. Comp. Sci. 1998, 38, 28.
|
| [36] |
Varamesh, A.; Hemmati-Sarapardeh, A.; Dabir, B.; Mohammadi, A. H. J. Mol. Liq. 2017, 242, 59.
doi: 10.1016/j.molliq.2017.06.039 |
| [37] |
Sola, D.; Ferri, A.; Banchero, M.; Manna, L.; Sicardi, S. Fluid Phase Equilib. 2008, 263, 33.
doi: 10.1016/j.fluid.2007.09.022 |
| [1] | 戚兴怡, 胡耀峰, 王若愚, 杨雅清, 赵宇飞. 机器学习在新材料筛选方面的应用进展[J]. 化学学报, 2023, 81(2): 158-174. |
| [2] | 韩逸之, 蓝建慧, 刘学, 石伟群. 基于机器学习势函数的熔盐体系分子动力学研究进展[J]. 化学学报, 2023, 81(11): 1663-1672. |
| [3] | 程敏, 王诗慧, 罗磊, 周利, 毕可鑫, 戴一阳, 吉旭. 面向乙烷/乙烯分离的金属有机框架膜的大规模计算筛选[J]. 化学学报, 2022, 80(9): 1277-1288. |
| [4] | 王诗慧, 薛小雨, 程敏, 陈少臣, 刘冲, 周利, 毕可鑫, 吉旭. 机器学习与分子模拟协同的CH4/H2分离金属有机框架高通量计算筛选[J]. 化学学报, 2022, 80(5): 614-624. |
| [5] | 蔡铖智, 李丽凤, 邓小梅, 李树华, 梁红, 乔智威. 基于机器学习和高通量计算筛选金属有机框架的甲烷/乙烷/丙烷分离性能[J]. 化学学报, 2020, 78(5): 427-436. |
| [6] | 朱博阳, 吴睿龙, 于曦. 人工智能助力当代化学研究[J]. 化学学报, 2020, 78(12): 1366-1382. |
| [7] | 刘治鲁, 李炜, 刘昊, 庄旭东, 李松. 金属有机骨架的高通量计算筛选研究进展[J]. 化学学报, 2019, 77(4): 323-339. |
| [8] | 谢江安, 梅虎, 吕娟, 潘显超, 王青, 张亚兰. 基于VHSE 结构表征的蛋白酶体酶切位点预测及酶切特异性研究[J]. 化学学报, 2012, 70(03): 318-324. |
| [9] | 禹新良. 自由基共聚合中单体Q-e活性参数预测[J]. 化学学报, 2010, 68(22): 2264-2272. |
| [10] | 刘东,章文军,许禄. 手性羟酸和氨基酸类化合物的构效关系研究[J]. 化学学报, 2009, 67(2): 145-150. |
| [11] | 包鑫,戴连奎. 加权最小二乘支持向量机稳健化迭代算法及其在光谱分析中的应用[J]. 化学学报, 2009, 67(10): 1081-1086. |
| [12] | 乔澍,谢昆,付川,祁俊生. 芳香胺类化合物N—H键离解能的定量构效关系研究[J]. 化学学报, 2009, 67(10): 1109-1115. |
| [13] | 刘涛,宋哲,焦春波,刘伟,朱鸣华,王晓钢. 基于SVM方法分析蛋白酶体裂解位点的特异性[J]. 化学学报, 2008, 66(21): 2341-2347. |
| [14] | 饶含兵, 李泽荣, 陈晓梅, 李象远. 基于支持向量学习机的HIV-1蛋白酶抑制剂的活性预测[J]. 化学学报, 2007, 65(3): 197-202. |
| [15] | 程存归,田玉梅,金文英. 应用小波特征提取肺癌组织FTIR的支持向量机分类方法研究[J]. 化学学报, 2007, 65(22): 2539-2543. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||