

β发卡多肽Trpzip4折叠的副本交换分子动力学模拟
收稿日期: 2013-01-04
网络出版日期: 2013-03-01
基金资助
项目受教育部新世纪优秀人才支持计划(No. NCET-07-0313)、国家自然科学基金(No. 20876052)及广东省自然科学基金(No. S2011010002078)资助.
Replica Exchange Molecular Dynamics Simulations on the Folding of Trpzip4 β-Hairpin
Received date: 2013-01-04
Online published: 2013-03-01
Supported by
Project supported by the Program for New Century Excellent Talents in Universities, Ministry of Education, China (No. NCET-07-0313), the National Natural Science Foundation of China (No. 20876052) and the Natural Science Foundation of Guangdong Province(No. S2011010002078).
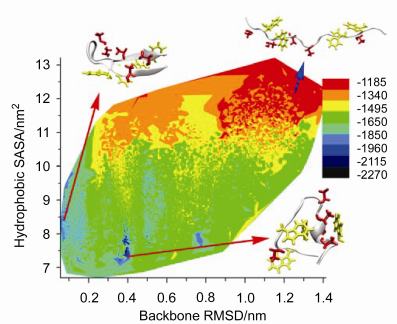
采用副本交换分子动力学对β发卡Trpzip4重折叠进行研究, 结果表明, 构象空间存在较大能垒时, 副本交换分子动力学(REMD)表现出比分子动力学(MD)更优的抽样效率. 288 K下, Trpzip4势能、骨架均方根偏差、溶剂可及表面积(SASA)在REMD中呈逐渐降低趋势. 采样得到两种特定形态的低势能构象: β发卡和螺旋-卷曲. β发卡中, 第3组氢键Asp46容易与Thr49形成氢键; 转角Thr49的羟基(OH)倾向于与Asp46的羧基(COO-)或主链上的C=O形成氢键, 强化了Asp46与Thr49的相互作用, 导致转角形成折回弯度; 与Thr49相比, Thr51的羟基倾向与水分子作用, 甲基朝内, 因此与Asp46的侧链作用微小. 螺旋-卷曲中, 吲哚环-甲基为主要疏水形式. Trpzip4在折叠成β发卡过程中, 转角氢键影响整个β发卡构象的形成. 转角氢键具有距离优势和强侧链相互作用, 最先形成.
廖晨伊 , 周健 . β发卡多肽Trpzip4折叠的副本交换分子动力学模拟[J]. 化学学报, 2013 , 71(04) : 593 -601 . DOI: 10.6023/A13010015
β-Hairpin is an essential secondary structure unit of protein. Understanding the formation mechanism of the β hairpin and kinetic stability helps to gain insight into the formation of protein secondary structure as well as the mechanism of protein folding. Replica exchange molecular dynamics (REMD) approach is applied to investigate the folding mechanism of Trpzip4 β-hairpin. The REMD method is more efficient than conventional MD in searching for a representative set of the low energy minima in the existence of a large energy gap between the native state and any of the other possible state. The potential energy, backbone RMSD and solvent-accessible surface area (SASA) present descending trends in the process of REMD at 288 K. Methyl groups (most in threonines) show hydrophobic interactions with SASA decrements. The hydrophobic interactions and intra hydrogen bond forming drive the folding and maintain the low energy structure. Two low energy structures, β-hairpin and helix-coil conformation, are sampled by REMD. In β-hairpin structure, the hydroxyl group (OH) of Thr49 at β turn is accessible to form hydrogen bonds with carboxyl group (COO-) or C=O group of Asp46; while the hydroxyl group (OH) of Thr51 is inclined to interact with water molecule with methyl orienting inwards. As a result, the strong interactions between Asp46 and Thr49 cause a bend at β turn. In helix-coil conformation, hydrophobic interactions play between indole ring and methyl group. One of the zip-in paths of Trpzip4 indicates that, the forming of hydrogen bonds at β turn affects the whole folding process of β hairpin. The forming of hydrogen bonds at β turn takes place first, since it has advantages of distance and strong interactions in both side chains and backbone. Total solvent-accessible surface area decreases significantly as it folds to the β hairpin structure. REMD method could sample large phase space for the low energy minima, and present scenarios of potential energy distribution on phase space with related variables as hydrophobic SASA, backbone RMSD, and hydrogen bonds for specific conformation. This work sheds some lights on the understanding of β hairpin folding.

/
| 〈 |
|
〉 |