

石墨烯纳米孔的制备及λ-DNA穿孔初步研究
收稿日期: 2013-12-03
网络出版日期: 2014-02-11
基金资助
项目受国家“863”计划(No. 2012AA02A104);自然科学基金(No. 51302037);上海交通大学重大项目及创新团队培育基金(No. X198775)和上海市重点学科建设基金(No. B209)资助.
Fabrication of Graphene Nanopores and a Preliminary Study on λ-DNA Translocation
Received date: 2013-12-03
Online published: 2014-02-11
Supported by
Project supported by the National High Technology Research and Development Program of China (No. 2012AA02A104), the National Natural Science Foundation of China (No. 51302037), the Innovative Research Team fund of Shanghai Jiaotong University (No. X198775) and the Shanghai Key Discipline fund (No. B209).
王跃 , 余旭丰 , 刘芸芸 , 谢骁 , 程秀兰 , 黄少铭 , 王志民 . 石墨烯纳米孔的制备及λ-DNA穿孔初步研究[J]. 化学学报, 2014 , 72(3) : 378 -381 . DOI: 10.6023/A13121208
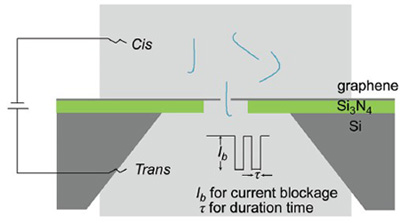
Nanopore sequencing is among the most promising technologies to achieve the goals of the "$1,000 Genome". Two major types of nanopores have been extensively investigated, protein and silicon-based solid-state nanopores. However, protein pores are short-lived and the length of solid-state nanopores is much larger than the distance between adjacent bases, resulting in incapability to discriminate individual bases along the single-stranded DNA molecules. In this paper, we report λ-DNA translocations through graphene nanopore. A large nanopore with diameter of about 30 nm on silicon nitride substrates were first fabricated using focused ion beam (FIB) system under the beam current of 2 pA, accelerating voltage of 30 kV and sculpting time of 1 s. Individual graphene membranes were suspended onto the substrates to cover the large pore, and nanopores with a diameter less than 10 nm are sculpted in the graphene sheet by focused electron beam (FEB) from a transmission electron microscope (TEM) under a 400 kx magnification times and 300 kV accelerating voltage at 1×105~5×105 A/m2 current density for 2~3 min. The edges of these graphene nanopores became smoother and sharper when the temperature was increased to about 450 ℃, which might help to lower interactions between graphene nanopores and the analytes. The signals of DNA translocation through graphene nanopores had been recorded using a patch clamp amplifier at 10 kHz sampling frequency filtered at 5 kHz via an integrated four-pole low-pass Bessel filter. Analyses of the DNA translocation current traces indicated that different conformations of DNA molecules may exist during entrance into nanopores. In addition, our overall detection platform had a low noise amplitude of around 10 pA, which allowed more sensitive signal detection. Taken together, our observations demonstrate that graphene nanopores are feasible for DNA sensing, leading a forward step towards single-molecule DNA sequencing using monolayered graphene nanopores.
Key words: graphene; nanopore; sequencing; focused ion beam; focused electron beam
[1] Bayley, H. Curr. Opin. Chem. Biol. 2006, 10, 628.
[2] Meyerson, M.; Gabriel, S.; Getz, G. Nat. Rev. Genet. 2010, 11, 685.
[3] Hayden, E. C. Nature 2008, 451, 378.
[4] Kayser, M.; de Knijff, P. Nat. Rev. Genet. 2011, 12, 179.
[5] Zhou, X.; Ren, L.; Li, Y.; Zhang, M.; Yu, Y.; Yu, J. Sci. China, Life Sci. 2010, 53, 44.
[6] Wanunu, M. Phys. Life Rev. 2012, 9, 125.
[7] Church, G.; Deamer, D. W.; Branton, D.; Baldarelli, R.; Kasianowicz, J. US 5795782, 1998 [Chem. Abstr. 1996, 126, 4207].
[8] Kasianowicz, J. J.; Brandin, E.; Branton, D.; Deamer, D. W. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13770.
[9] Schneider, G. F.; Dekker, C. Nat. Biotech. 2012, 30, 326.
[10] Venkatesan, B. M.; Bashir, R. Nat. Nanotech. 2011, 6, 615.
[11] Deamer, D. W.; Akeson, M. Trends Biotech. 2000, 18, 147.
[12] Li, J.; Stein, D.; McMullan, C.; Branton, D.; Aziz, M. J.; Golovchenko, J. A. Nature 2001, 412, 166.
[13] Storm, A. J.; Chen, J. H.; Ling, X. S.; Zandbergen, H. W.; Dekker, C. Nat. Mater. 2003, 2, 537.
[14] Siwy, Z.; Fuliński, A. Phys. Rev. Lett. 2002, 89, 198103.
[15] Langecker, M.; Arnaut, V.; Martin, T. G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F. C. Science 2012, 338, 932.
[16] Liu, H.; He, J.; Tang, J.; Liu, H.; Pang, P.; Cao, D.; Krstic, P.; Joseph, S.; Lindsay, S.; Nuckolls, C. Science 2010, 327, 64.
[17] Wang, Z.; Huang, S. CN ZL200610147236. 3, 2006[Chem. Abstr. 2007, 147, 230108].
[18] Sha, J.; Hasan, T.; Milana, S.; Bertulli, C.; Bell, N. A. W.; Privitera, G.; Ni, Z.; Chen, Y.; Bonaccorso, F.; Ferrari, A. C.; Keyser, U. F.; Huang, Y. Y. S. ACS Nano 2013, 7, 8857.
[19] Liu, S.; Lu, B.; Zhao, Q.; Li, J.; Gao, T.; Chen, Y.; Zhang, Y.; Liu, Z.; Fan, Z.; Yang, F.; You, L.; Yu, D. Adv. Mater. 2013, 25, 4549.
[20] Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.; Branton, D. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 1079.
[21] Venta, K.; Shemer, G.; Puster, M.; Rodriguez-Manzo, J. A.; Balan, A.; Rosenstein, J. K.; Shepard, K.; Drndi?, M. ACS Nano 2013, 7, 4629.
[22] Ying, Y.-L.; Zhang, J.; Gao, R.; Long, Y.-T. Angew. Chem., Int. Ed. 2013, 52, 13154.
[23] Ying, Y.; Zhang, X.; Liu, Y.; Xue, M.; Li, H.; Long, Y. Acta Chim. Sinica 2013, 71, 44. (应佚伦, 张星, 刘钰, 薛梦竹, 李洪林, 龙亿涛, 化学学报, 2013, 71, 44.)
[24] Venta, K.; Wanunu, M.; Drndi?, M. Nano Lett. 2013, 13, 423.
[25] Liu, A.; Zhao, Q.; Guan, X. Anal. Chim. Acta 2010, 675, 106.
[26] Garaj, S.; Hubbard, W.; Reina, A.; Kong, J.; Branton, D.; Golovchenko, J. Nature 2010, 467, 190.
[27] Merchant, C. A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fischbein, M. D.; Venta, K.; Luo, Z.; Johnson, A. T. C.; Drndi?, M. Nano Lett. 2010, 10, 2915.
[28] Nelson, T.; Zhang, B.; Prezhdo, O. V. Nano Lett. 2010, 10, 3237.
[29] Postma, H. W. Nano Lett. 2010, 10, 420.
[30] Schneider, G. F.; Kowalczyk, S. W.; Calado, V. E.; Pandraud, G.; Zandbergen, H. W.; Vandersypen, L. M.; Dekker, C. Nano Lett. 2010, 10, 3163.
[31] Kim, M. J.; McNally, B.; Murata, K.; Meller, A. Nanotechnology 2007, 18, 205302.
[32] Liu, Q.; Wu, H.; Wu, L.; Xie, X.; Kong, J.; Ye, X.; Liu, L. Plos One 2012, 7, e46014.
[33] Wanunu, M.; Sutin, J.; McNally, B.; Chow, A.; Meller, A. Biophys. J. 2008, 95, 4716.
[34] Fologea, D.; Gershow, M.; Ledden, B.; McNabb, D. S.; Golovchenko, J. A.; Li, J. Nano Lett. 2005, 5, 1905.
[35] Tabard-Cossa, V.; Trivedi, D.; Wiggin, M.; Jetha, N. N.; Marziali, A. Nanotechnology 2007, 18, 305505.
[36] Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Nat. Nanotech. 2009, 4, 265.
[37] Li, J.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J. A. Nat. Mater. 2003, 2, 611.
[38] Smeets, R. M. M.; Keyser, U. F.; Dekker, N. H.; Dekker, C. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 417.
[39] Hoogerheide, D. P.; Garaj, S.; Golovchenko, J. A. Phys. Rev. Lett. 2009, 102, 256804.
[40] Bayley, H. Nature 2010, 467, 164.
/
| 〈 |
|
〉 |