

混合模式色谱材料Click TE-GSH高效富集多磷酸化肽
收稿日期: 2014-07-22
修回日期: 2014-08-25
网络出版日期: 2014-08-25
基金资助
项目受国家自然科学基金(Nos. 81171486, 21105100)和辽宁省特聘教授资助.
Highly Selective Enrichment of Multi-Phosphopeptides by Click TE-GSH
Received date: 2014-07-22
Revised date: 2014-08-25
Online published: 2014-08-25
Supported by
Project supported by the National Natural Science Foundation of China (Nos. 81171486, 21105100) and Distinguished Professor of Liaoning Province.
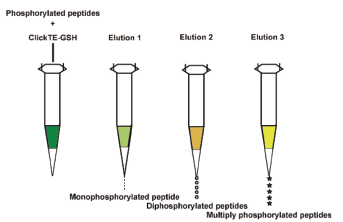
多磷酸蛋白对于生物体适应内外环境具有重要意义,而明确多磷酸蛋白的磷酸位点功能及其信号转导机制尤为关键. 复杂生物样品中多磷酸化肽的低丰度、低电离的特性,以及非磷酸化肽的抑制作用,决定了质谱分析前进行多磷酸化肽富集是非常必要的步骤. 本工作采用基于巯基-烯烃点击化学法合成的混合模式材料Click TE-GSH进行单磷酸化肽和多磷酸化肽的选择性富集. 我们建立了单磷酸化肽、双磷酸化肽和多磷酸化肽的顺序分段富集方法. 该优化方法能抗干扰,应用于脱脂牛奶时富集到11条多磷酸化肽. 与商品化固化金属亲和色谱(IMAC)材料相比,Click TE-GSH富集多磷酸化肽的选择性更好. 本工作所建立的富集方法为高效富集多磷酸化肽提供新方法和新技术.
关键词: 多磷酸化肽; Click TE-GSH; 高效富集; 质谱
丰小敏 , 沈爱金 , 李先琴 , 李秀玲 , 邹丽娟 , 梁鑫淼 . 混合模式色谱材料Click TE-GSH高效富集多磷酸化肽[J]. 化学学报, 2014 , 72(10) : 1085 -1091 . DOI: 10.6023/A14070546
Multisite phosphorylation of proteins plays an important role in signal transduction. However, it is difficult to describe precisely the mechanism of these phosphorylation cascades. In order to study the function of phosphorylated proteins in a deep-going way, enrichment protocol with high selectivity and coverage is urgent needed. In the present study Click TE-GSH, a novel HILIC material synthetized in our group, was used to enrich and sequentially elute phosphorylated peptides in different fractions. As a mixed-mode chromatographic material, Click TE-GSH exhibits both hydrophilic interaction and cation-exchange characteristics. To understand the retention mechanism, we firstly carried out investigation to study the influence of acetonitrile (ACN) concentration, solution pH value and salt concentration on the retention of phosphorylated peptides respectively. The results showed that phosphorylated peptides were eluted with low concentration of ACN and non-phosphorylated peptides were eluted with high concentration of ACN, which was in accordance with the hydrophilic retention characteristics of Click TE-GSH material. Meanwhile, pH value and salt concentration affected the retention of phosphorylated peptides, owing to the change of surface charge on the stationary phase. Under the optimized condition, mono-phosphorylated peptide, di-phosphorylated peptides and multiply phosphorylated peptides were enriched selectively. 6 mono-phosphorylated peptides, 2 di-phosphorylated peptides and 15 multiply phosphorylated peptides were effectively enriched and detected. By contrast, immobilized metal ion affinity chromatography (IMAC) was utilized to enrich phosphorylated peptides. 2 mono-phosphorylated peptides and 6 multiply phosphorylated peptides were identified. It is obvious the enrichment efficiency of Click TE-GSH was much higher than that of IMAC. The established method was validated with relatively complex sample, including peptide mixture of α-casein and bovine serum albumin (BSA) at the molar ratio of 1:1 and 1:10. In addition, Click TE-GSH was applied to enrich phosphorylated peptides in milk. 11 multiply phosphorylated peptides were successfully identified. The result proved the excellent selectivity of the method. Therefore, this optimized protocol has great potential for the enrichment of multiply phosphorylated peptides.

[1] Olsen, J. V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Cell 2006, 127, 635.
[2] (a) Cohen, P. Eur. J. Biochem. 2001, 268, 5001.
(b) Huang, O. W.; Ma, X. L.; Yin, J. P.; Flinders, J.; Maurer, T.; Kayagaki, N.; Phung, Q.; Bosanac, I.; Arnott, D.; Dixit, V. M.; Hymowitz, S. G.; Starovasnik, M. A.; Cochran, A. G. Nat. Struct. Mol. Biol. 2012, 19, 171.
[3] Vicente-Manzanares, M.; Horwitz, A. R. Biochem. Biophys. Res. Commun. 2010, 402, 537.
[4] Deneubourg, L.; Edimo, W. E.; Moreau, C.; Vanderwinden, J.-M.; Erneux, C. Cell Signal. 2014, 26, 1193.
[5] Sims, R. J.; Reinberg, D. Nat. Rev. Mol. Cell Biol. 2008, 9, 815.
[6] Zou, Y.; Jiang, W. H.; Zou, L. J.; Li, X. L.; Liang, X. M. Chin. J. Chromatogr. 2013, 31, 367. (邹瑶, 姜武辉, 邹丽娟, 李秀玲, 梁鑫淼, 色谱, 2013, 31, 367.)
[7] Bai, Z. F.; Wang, H. X. Chinese J. Anal. Chem. 2009, 37, 1382. (柏兆方, 王红霞, 分析化学, 2009, 37, 1382.)
[8] Sui, S. H.; Wang, J. L.; Lu, Z.; Cai, Y.; Zhang, Y. J.; Yu, W. F.; Qian, X. H. Chin. J. Chromatogr. 2008, 26, 195. (隋少卉, 王京兰, 卢庄, 蔡耘, 张养军, 蔚文峰, 钱小红, 色谱, 2008, 26, 195.)
[9] Li, X. L.; Guo, Z. M.; Sheng, Q. Y.; Xue, X. Y.; Liang, X. M. Analyst 2012, 137, 2774.
[10] Li, S. H.; Dass, C. Anal. Biochem. 1999, 270, 9.
[11] Larsen, M. R.; Thingholm, T. E.; Jensen, O. N.; Roepstorff, P.; Jorgensen, T. J. D. Mol. Cell. Proteomics 2005, 4, 873.
[12] Thingholm, T. E.; Jensen, O. N.; Robinson, P. J.; Larsen, M. R. Mol. Cell. Proteomics 2008, 7, 661.
[13] Zhong, H. Y.; Xiao, X.; Zheng, S.; Zhang, W. Y.; Ding, M. J.; Jiang, H. Y.; Huang, L. L.; Kang, J. Nat. Commun. 2013, 4, 1656.
[14] (a) McNulty, D. E.; Annan, R. S. Mol. Cell. Proteomics 2008, 7, 971.
(b) Albuquerque, C. P.; Smolka, M. B.; Payne, S. H.; Bafna, V.; Eng, J.; Zhou, H. L. Mol. Cell. Proteomics 2008, 7, 1389.
[15] Shen, A. J.; Guo, Z. M.; Liang, X. M. Prog. Chem. 2014, 26, 10. (沈爱金, 郭志谋, 梁鑫淼, 化学进展, 2014, 26, 10.)
[16] Ke, C. Y.; Sun, W. J.; Zhang, Q. Z.; Zheng, L. Acta Chim. Sinica 2012, 70, 1637. (柯从玉, 孙妩娟, 张群正, 郑莉, 化学学报, 2012, 70, 1637.)
[17] Shen, A. J.; Li, X. L.; Dong, X. F.; Wei, J.; Guo, Z. M.; Liang, X. M. J. Chromatogr. A 2013, 1314, 63.
/
| 〈 |
|
〉 |