

亚硫酸钠介导的s4U转化检测转录组新生RNA中m6A
收稿日期: 2020-10-15
网络出版日期: 2020-11-27
基金资助
项目受国家自然科学基金(91753201); 项目受国家自然科学基金(21721005); 项目受国家自然科学基金(21907079)
Sodium Sulfite-Mediated s4U Conversion for m6A Profiling of Nascent Transcriptome-Wide RNA
Received date: 2020-10-15
Online published: 2020-11-27
Supported by
National Natural Science Foundation of China(91753201); National Natural Science Foundation of China(21721005); National Natural Science Foundation of China(21907079)
危琦 , 韩少卿 , 陈玉琪 , 周翔 . 亚硫酸钠介导的s4U转化检测转录组新生RNA中m6A[J]. 化学学报, 2021 , 79(3) : 326 -330 . DOI: 10.6023/A20100477
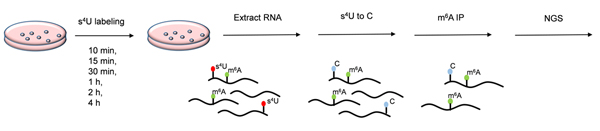
RNA sequencing can profile gene expression at steady-state level but is weak in studying temporal RNA dynamics. A common method for nascent RNA analysis is metabolic labeling with nucleoside analog. We developed a method for nascent RNA sequencing based on sodium sulfite-mediated 4-thiouridine-to-cytidine (s4U-to-C) conversion. The RNA contained s4U is reacted in the buffer (10 mmol/L Na2SO3, 200 mmol/L NH4Cl and 200 mmol/L Na2HPO4/NaH2PO4, pH 7.4) at 70 ℃ for 4 h, the yield can reach more than 90%. This method can efficiently distinguish nascent RNA information from total RNAs. SUC-seq is used to investigate the m6A, which is the most prevalent modification in mammalian mRNA and has been shown to have an essential regulatory role in the control of gene expression. Here, we provide a time resolved high-resolution profiling of m6A on nascent RNA transcripts. 3×106 HEK293T cells were seeded per 10-cm cell dish and grown for 24 h. Then, s4U was added to the medium at a final concertation of 500 μmol/L chased for 0, 10, 15, 30, 60, 120 and 240 min. Total RNA was extracted using TRIzol reagent. 10 μg total RNA was fragmented using RNA fragmentation reagents at 70 ℃ for 10 min after gDNA was removed. Fragmented RNA was then subjected to s 4U reaction to realize the conversion of s4U to C. Subsequently, 5 μg treated RNA were taken to perform m 6A immunoprecipitation. After removing rRNA, library was constructed and perform next-generation sequencing. A large number of T to C mutations were observed in the mapping reads. The T to C mutation rate of the control RNA was kept at a low background level. The results show that our method can successfully distinguish nascent RNA from total RNA. Via analyzing the nascent RNA in m6A, the nascent m6A can be distinguished from the total m6A. This method can be a promising strategy to study the mechanism and function of m6A.
Key words: sodium sulfite; nascent RNA; 4-thiouridine; N6-methyladenosine
| [1] | Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; Zhao, X.; Li, A.; Yang, Y.; Dahal, U.; Lou, X.-M.; Liu, X.; Huang, J.; Yuan, W.-P.; Zhu, X.-F.; Cheng, T.; Zhao, Y.-L.; Wang, X.; Danielsen, J. M. R.; Liu, F.; Yang, Y.-G. Cell Res. 2014, 24,177. |
| [2] | Frye, M.; Harada, B. T.; Behm, M.; He, C. Science 2018, 361,1346. |
| [3] | Roundtree, I. A.; He, C. Curr. Opin. Chem. Biol. 2016, 30,46. |
| [4] | Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Nat. Rev. Genet. 2014, 15,293. |
| [5] | Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G. Nat. Chem. Biol. 2011, 7,885. |
| [6] | Gasser, C.; Delazer, I.; Neuner, E.; Pascher, K.; Brillet, K.; Klotz, S.; Trixl, L.; Himmelstoss, M.; Ennifar, E.; Rieder, D.; Lusser, A.; Micura, R. Angew. Chem. Int. Ed. 2020, 59,6881. |
| [7] | Herzog, V. A.; Reichholf, B.; Neumann, T.; Rescheneder, P.; Bhat, P.; Burkard, T. R.; Wlotzka, W.; von Haeseler, A.; Zuber, J.; Ameres, S. L. Nat. Methods 2017, 14,1198. |
| [8] | Schofield, J. A.; Duffy, E. E.; Kiefer, L.; Sullivan, M. C.; Simon, M. D. Nat. Methods 2018, 15,221. |
| [9] | Riml, C.; Amort, T.; Rieder, D.; Gasser, C.; Lusser, A.; Micura, R. Angew. Chem. Int. Ed. 2017, 56,13479. |
| [10] | Chen, Y.; Wu, F.; Chen, Z.; He, Z.; Wei, Q.; Zeng, W.; Chen, K.; Xiao, F.; Yuan, Y.; Weng, X.; Zhou, Y.; Zhou, X. Adv. Sci. 2020, 7,1900997. |
/
| 〈 |
|
〉 |