1 引言
2 结果与讨论
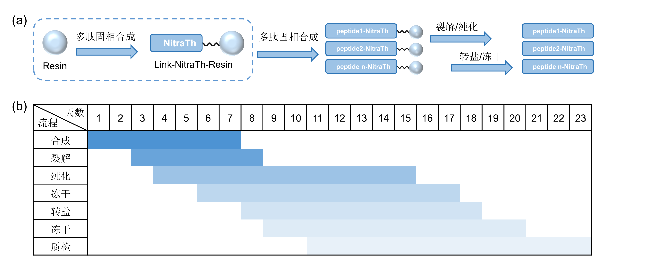
2.1 多肽制备流程
2.2 通用肽段Link-NitraTh的合成方法优化
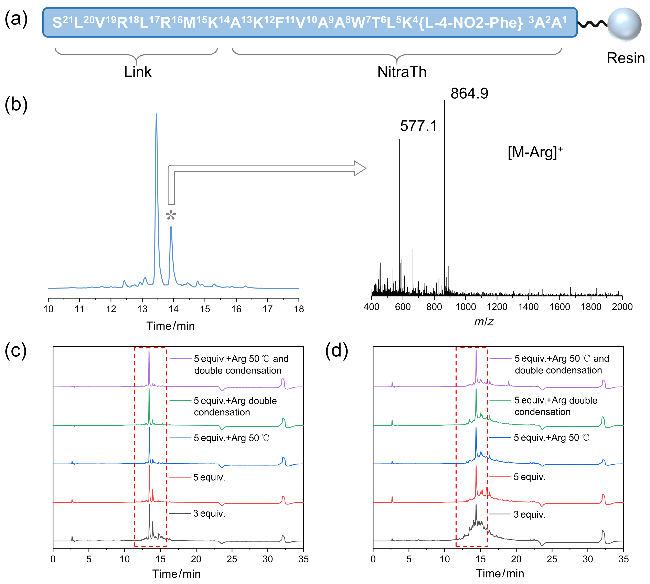
图2 (a) Link-NitraTh多肽树脂示意图; (b)氨基酸5 equiv.合成N16的HPLC及主要杂质分析图; (c)不同条件下制备得到的N16多肽HPLC分析图; (d)不同条件下制备得到的Link-NitraTh多肽HPLC分析图Figure 2 (a) Schematic diagram of Link-NitraTh peptide resin; (b) HPLC diagram and main impurity analysis diagram of N16 with 5 equiv. amino acid; (c) HPLC diagram of N16 peptides prepared under different conditions; and (d) HPLC diagram of Link-NitraTh peptides prepared under different conditions |
表1 不同条件下Link-NitraTh片段的制备结果Table 1 Results of the Link-NitraTh fragment production under different conditions |
| 多肽片段 | n(氨基酸)∶ n(树脂) | 50 ℃(Arg) | 双缩(Arg) | 纯度/% | |
|---|---|---|---|---|---|
| N6 | 3 | — | — | 79.86 | |
| N6 | 5 | — | — | 90.61 | |
| N10 | 3 | — | — | 49.61 | |
| N10 | 5 | — | — | 90.54 | |
| N16 | 3 | 否 | 否 | 28.31 | |
| N16 | 5 | 否 | 否 | 54.04 | |
| N16 | 5 | 是 | 否 | 67.63 | |
| N16 | 5 | 否 | 是 | 65.52 | |
| N16 | 5 | 是 | 是 | 73.19 | |
| Link-NitraTh | 3 | 否 | 否 | 9.12 | |
| Link-NitraTh | 5 | 否 | 否 | 21.50 | |
| Link-NitraTh | 5 | 是 | 否 | 23.03 | |
| Link-NitraTh | 5 | 否 | 是 | 23.09 | |
| Link-NitraTh | 5 | 是 | 是 | 33.54 | |
2.3 新生抗原多肽制备实例
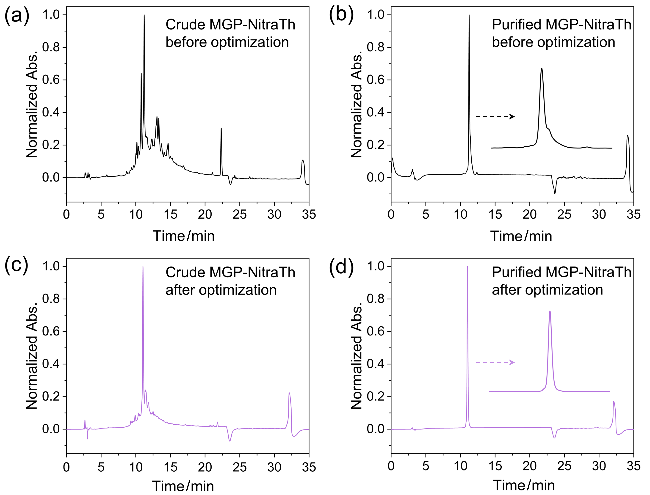
图3 (a)优化前合成MGP-NitraTh的粗品HPLC分析图; (b)优化前合成MGP-NitraTh的精品HPLC分析图; (c)优化后合成MGP-NitraTh的粗品HPLC分析图; (d)优化后合成MGP-NitraTh的精品HPLC分析图.Figure 3 (a) The HPLC chromatogram of crude MGP-NitraTh before optimization; (b) The HPLC chromatogram of purified MGP-NitraTh before optimization; (c) The HPLC chromatogram of crude MGP-NitraTh after optimization; (d) The HPLC chromatogram of purified MGP-NitraTh after optimization |
表2 新生抗原多肽制备实例Table 2 Neoantigen peptide preparation examples |
| 序列组 | 样品名 | 序列(38AA) | 外购纯度/% | 自制纯度/% | ||
|---|---|---|---|---|---|---|
| NJ81-01 | SLC38A4-NitraTh | SVGIILPLPLLKNLGYLSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | 94.98 | 97.60 | ||
| CTNNB1-NitraTh | SGATTTAPPLSGKGNPESLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | 99.86 | 96.15 | |||
| VIL1-NitraTh | KLYHVSDSKGNLVVREVSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | 93.40 | 98.23 | |||
| PSME4-NitraTh | LQQSKNPSVNQILLSPESLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | 100 | 97.02 | |||
| POC1B-NitraTh | NVLTQTVSVLEQRLTLTSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —a | —a | |||
| CDYL2-NitraTh | CGRKLTAQVACSRGLVSSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —a | —a | |||
| TRMT6-NitraTh | YDTLAQMLMLGNIRAGNSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | 93.76 | 98.77 | |||
| IFNAR2-NitraTh | PEVDVELPMMPKDSPQQSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 94.75 | |||
| KDR-NitraTh | SLVEATVGKRVRIPAKYSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 99.19 | |||
| NJ81-02 | NRCAM-NitraTh | NLSDTEFYVAKSSRERPSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 98.34 | ||
| MFSD5-NitraTh | PYLYKLYQRYYFLEGQISLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 95.70 | |||
| TMEM127-NitraTh | FAVSFYLVEGAGGASILSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | —a | |||
| BSDC1-NitraTh | NACWPHRRPRAQASSQSSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 97.53 | |||
| ARHGAP25-NitraTh | EVLFPKSKNIPLSPPAQSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 95.03 | |||
| PCNX1-NitraTh | HLTRSLQHILCGDLLLGSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | 95.54 | |||
| CDC42BPB-NitraTh | FLSQQSFDACAVELESSLVRLRMKAKFVAAWTLK{L-4-NO2-Phe}AA | —b | —a | |||
a The peptide was obtained because of poor solubility; b peptide was not prepared. |