

Rif-orf13编码的细胞色素P450催化利福霉素生物合成过程中C34a位的羟化反应
收稿日期: 2018-11-12
修回日期: 2018-12-06
网络出版日期: 2018-12-17
34a-Hydroxylation in Rifamycin Biosynthesis Catalyzed by Cytochrome P450 Encoded by rif-orf13
Received date: 2018-11-12
Revised date: 2018-12-06
Online published: 2018-12-17
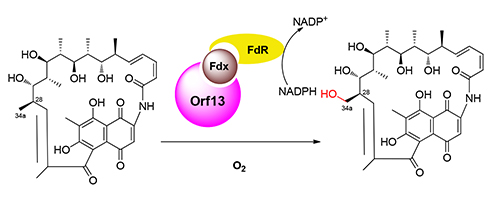
利福霉素生物合成途径在经历了二十余年的研究之后,仍然没有得到完全阐明.其中C34a甲基的氧化脱除是利福霉素成熟过程中的必需反应步骤,但是催化这一步骤的酶尚未鉴定·推测可能是利福霉素生物合成基因簇编码的某个细胞色素P450催化了这一步骤.选取利福霉素生物合成基因簇中功能尚未确证的P450基因rif-orf0、rif-orf4和rif-orf13在变铅青链霉菌中进行异源表达和底物喂养实验,发现表达了rif-orf13的链霉菌能够将16-脱甲基-34a-脱氧利福霉素W (1)转化为16-脱甲基利福霉素W (2).将rif-orf13在大肠杆菌BL21(DE3)中进行诱导表达,利用纯化的Orf13蛋白进行体外酶催化反应,发现Orf13能够将底物1羟化为产物2.结合前人的基因敲除研究,认为rif-orf13是编码34a-脱氧利福霉素W羟化酶的基因,其在胞内的功能可以被另一个负责C12-C29双键氧化断裂的P450基因rif-orf5替代.
周强 , 罗光彩 , 张惠展 , 唐功利 . Rif-orf13编码的细胞色素P450催化利福霉素生物合成过程中C34a位的羟化反应[J]. 有机化学, 2019 , 39(4) : 1169 -1174 . DOI: 10.6023/cjoc201811018
The biosynthetic pathway of rifamycins is still not completely deciphered after decades of study. For example, the gene responsible for the oxidative elimination of C34a is not identified. It was proposed that some cytochrome P450 is related to this essential biosynthetic step in the modification of rifamycin. Here, cytochrome P450 encoding genes rif-orf0, 4 and 13 from rifamycin biosynthetic gene cluster were heterologously expressed in Streptomyces lividans and fed with 16-demethyl-34a-deoxyrifamycin W (1). Compound 1 was completely converted into 16-demethylrifamycin W (2) in the strain harboring rif-orf13. His6-tagged Orf13 was prepared from E. coli BL21 (DE3) and characterized to be active cytochrome P450. Enzymatic assays demonstrated that compound 1 could be converted into 2 by Orf13 as in vivo. Therefore, we concluded that rif-orf13 is responsible for the hydroxylation on C34a in the biosynthesis of rifamycins. In addition, it's role in vivo could be functionally complemented by rif-orf5, which encoding another cytochrome P450 enzyme.

Key words: cytochrome P450; hydroxylation; rifamycin; rif-orf13
[1] Schupp, T.; Toupet, C.; Engel, N.; Goff, S. FEMS Microbiol. Lett. 1998, 159, 201.
[2] August, P. R.; Tang, L.; Yoon, Y. J.; Ning, S.; Muller, R.; Yu, T. W.; Taylor, M.; Hoffmann, D.; Kim, C. G.; Zhang, X.; Hutchinson, C. R.; Floss, H. G. Chem. Biol. 1998, 5, 69.
[3] Yu, T. W.; Shen, Y.; Doi-Katayama, Y.; Tang, L.; Park, C.; Moore, B. S.; Hutchinson, C. R.; Floss, H. G. Proc. Natl. Acad Sci. U. S. A. 1999, 96, 9051.
[4] Wilson, M. C.; Gulder, T. A.; Mahmud, T.; Moore, B. S. J. Am. Chem. Soc. 2010, 132, 12757.
[5] Xu, J.; Wan, E.; Kim, C. J.; Floss, H. G.; Mahmud, T. Microbiology 2005, 151, 2515.
[6] Xu, J.; Mahmud, T.; Floss, H. G. Arch. Biochem. Biophys. 2003, 411, 277.
[7] Yuan, H.; Zhao, W.; Zhong, Y.; Wang, J.; Qin, Z.; Ding, X.; Zhao, G. P. Acta Biochim. Biophys. Sin. (Shanghai) 2011, 43, 948.
[8] Qi, F.; Lei, C.; Li, F.; Zhang, X.; Wang, J.; Zhang, W.; Fan, Z.; Li, W.; Tang, G. L.; Xiao, Y.; Zhao, G.; Li, S. Nat. Commun. 2018, 9, 2342.
[9] Campbell, E. A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S. A. Cell 2001, 104, 901.
[10] Floss, H. G.; Yu, T. W. Chem. Rev. 2005, 105, 621.
[11] Lee, S. K.; Choi, C. Y.; Ahn, J. S.; Cho, J. Y.; Park, C. S.; Yoon, Y. J. J. Microbiol. Biotechnol. 2004, 14, 356.
[12] Zhou, Q.; Luo, G.-C.; Zhang, H.; Tang, G.-L. Appl. Environ. Microbiol. 2018, DOI:10.1128/AEM.02597-18.
[13] Omura, T.; Sato, R. J. Biol. Chem. 1964, 239, 2370.
[14] Cook, D. J.; Finnigan, J. D.; Cook, K.; Black, G. W.; Charnock, S. J. Adv. Protein Chem. Struct. Biol. 2016, 105, 105.
[15] Li, S.; Podust, L. M.; Sherman, D. H. J. Am. Chem. Soc. 2007, 129, 12940.
[16] Zhang, W.; Liu, Y.; Yan, J.; Cao, S.; Bai, F.; Yang, Y.; Huang, S.; Yao, L.; Anzai, Y.; Kato, F.; Podust, L. M.; Sherman, D. H.; Li, S. J. Am. Chem. Soc. 2014, 136, 3640.
[17] Kishi, T.; Yamana, H.; Muroi, M.; Harada, S.; Asai, M. J. Antibiot. (Tokyo) 1972, 25, 11.
/
| 〈 |
|
〉 |