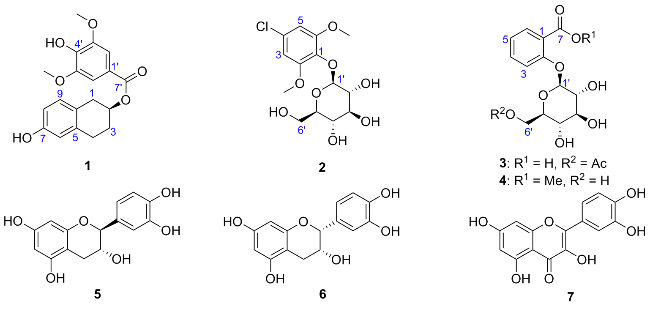
Compound
3 was obtained as white amorphous powder, with the molecular formula C
15H
18O
9 from HRESIMS
m/
z 341.0875 [M-H]
- (C
15H
17O
9, calcd 341.0878) combined with
1H NMR and
13C NMR spectroscopic data. In the
1H NMR spectrum, four aromatic protons at
δ 7.59 (d,
J=7.6 Hz, 1H, H-6), 7.48 (dd,
J=8.4, 7.6 Hz, 1H, H-4), 7.17 (d,
J=8.4 Hz, 1H, H-3) and 7.05 (dd,
J=7.6, 7.6 Hz, 1H, H-5), one singlet methyl group at
δ 1.96 (s, 3H, H-2''), together with many protons arising from sugar moiety were observed. The
13C NMR data showed 15 resonances, including two carbonyl carbons (
δ 170.7 and 167.6), six aromatic carbons (
δ 155.7, 133.7, 130.8, 122.3, 122.0 and 116.8), six
O-bearing carbons, and one methyl carbon (
δ 21.1), which are characteristic signals of a phenolic acid glycoside. The
1H NMR and
13C NMR data (
Table 1) were very similar to those of known compound
4, except for the presence of one acetyl group signal [
δH 1.96 (3H) and
δC 167.6/21.1] in
3 and the absence of a methoxyl group signal (
δC 52.0) in
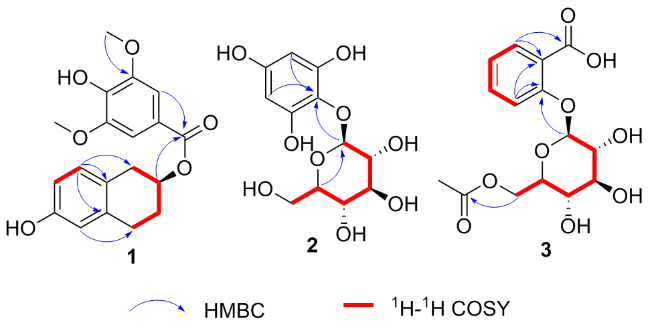
4. The location of the acetyl group at C-6' was confirmed by the HMBC correlations from H-6' to C-1'' (
Figure 2). The HMBC correlations from H-1' to C-2 indicated that C-1' linked with C-2 by a glycosidic bond. The HMBC correlations from H-6 to C-1/7 and H-3 to C-1 indicated that the carboxyl group located at C-1. Detailed analysis of 2D NMR (HSQC,
1H-
1H COSY and HMBC) spectra confirmed that the other part of the molecule was the same as those of
4. The glycone part of
3, a glucose moiety, was identified and characterized by the anomeric proton doublet at
δH 4.91 (d,
J=6.0 Hz, 1H, H-1')/
δc 101.1. These data suggested that the glycosyl moiety was
β- glucose. According to the biogenic synthesis pathway and the similar optical rotation value of compounds
3 and
4, the glycosyl moiety was assigned as
D-glucose. Furthermore, it was also determined by the PMP-labeling HPLC analysis method. The retention time of the PMP-labeling product of compound
3 (retention time: 44.87 min) was the same as the retention time of the PMP-labeling product of
D-glucose standard (retention time: 44.87 min). The optical rotation value of the glucose from compound
3 (
$[\alpha]_{\mathrm{D}}^{24}$+51.4, H
2O) was also the same as the standard
D-glucose (
$[\alpha]_{\mathrm{D}}^{24}$+52.0, H
2O). This result indicated that the glycosyl of compound
3 was
D-glucose (
Figure 3). Thus, compound
3 was identified as salicylic acid-2-
O-(6'-
O-acetyl)-
β-
D-glucopyranoside, a new phenolic glycoside.